Single-molecule tracking reveals dynamic regulation of ribosomal scanning
- PMID: 39356761
- PMCID: PMC11446271
- DOI: 10.1126/sciadv.adm9801
Single-molecule tracking reveals dynamic regulation of ribosomal scanning
Abstract
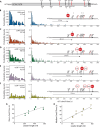
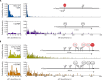
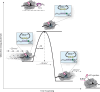
How eukaryotic ribosomes traverse messenger RNA (mRNA) leader sequences to search for protein-synthesis start sites remains one of the most mysterious aspects of translation and its regulation. While the search process is conventionally described by a linear "scanning" model, its exquisitely dynamic nature has restricted detailed mechanistic study. Here, we observed single Saccharomyces cerevisiae ribosomal scanning complexes in real time, finding that they scan diverse mRNA leaders at a rate of 10 to 20 nt s-1. We show that specific binding of a protein to its mRNA leader sequence substantially arrests scanning. Conversely, impairing scanning-complex guanosine 5'-triphosphate hydrolysis results in native start-site bypass. Our results illustrate an mRNA-centric, kinetically controlled regulatory model where the ribosomal pre-initiation complex amplifies a nuanced energetic landscape to regulate scanning and start-site selection fidelity.
Figures


References
-
- Aitken C. E., Lorsch J. R., A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19, 568–576 (2012). - PubMed
-
- Kozak M., How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell 15, 1109–1123 (1978). - PubMed
-
- Richardson J. P., Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta 1577, 251–260 (2002). - PubMed
-
- Berthelot K., Muldoon M., Rajkowitsch L., Hughes J., McCarthy J. E. G., Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol. Microbiol. 51, 987–1001 (2004). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

