Genomic and clinical landscape of metastatic hormone receptors-positive breast cancers carrying ESR1 alterations
- PMID: 39357123
- PMCID: PMC11480226
- DOI: 10.1016/j.esmoop.2024.103731
Genomic and clinical landscape of metastatic hormone receptors-positive breast cancers carrying ESR1 alterations
Abstract
Background: Somatic genetic alterations of the estrogen receptor 1 gene (ESR1) are enriched in endocrine therapy-resistant, estrogen receptor-positive (ER+) metastatic breast cancer (mBC). Herein, we investigated and compared the clinical and genomic landscape of ESR1-mutant (ESR1MUT) and ESR1 wild type (ESR1WT) ER+/ human epidermal growth factor receptor 2 (HER2)- mBCs.
Methods: Clinical and genomic data were retrieved from cBioPortal using the publicly-available MSK MetTropism dataset. Metastatic, ER+/HER2- mBC samples were included in the analysis. Only oncogenic and likely oncogenic alterations according to OncoKB were included. Statistical analyses were carried out using alpha level of 0.05, with a false discovery rate threshold of 10% for multiple comparisons using the Benjamini-Hochberg method.
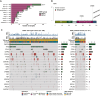
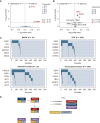
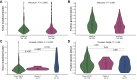
Results: Among 679 samples, 136 ESR1MUT among 131 tumors were found (19.2%). The frequency of ESR1MUT was higher in ductal versus lobular mBC (21.2% versus 13.8%, P = 0.052) and enriched in liver metastasis compared with other sites (22.5% versus 12.7%; q = 0.02). Compared with ESR1WT mBC, ESR1MUT tumors showed higher fraction of genome altered (FGA) {[0.28 interquartile range (IQR), 0.15-0.43] versus 0.22 (0.11-0.38); P = 0.04} and tumor mutational burden (TMB) [4.89 (IQR 3.46-6.85) versus 3.92 (2.59-6.05) mut/Mb; P = 0.001]. Tumors harboring p.E380X alterations showed higher TMB compared with those with H11-12 alterations [8.24 (IQR 5.06-15.3) versus 4.89 (IQR 3.46-6.75) mut/Mb; P = 0.01]. Genetic alterations of TP53 were enriched in ESR1WT tumors (36% versus 14%) [odds ratio (OR) 3.17, 95% confidence interval (CI) 1.88-5.64, q = 0.001]. Considering signaling pathways, ESR1MUT tumors showed a lower occurrence of TP53 (OR 0.48, 95% CI 0.30-0.74; q = 0.003) and MAPK (OR 0.29, 95% CI 0.11-0.65; q = 0.009) alterations. TP53 (q < 0.001), CDH1 (q < 0.001), and ERBB2 (q < 0.001) demonstrated mutual exclusivity with ESR1MUT.
Conclusions: ER+/HER2- mBCs carrying ESR1MUT exhibit a divergent genomic background, characterized by a lower prevalence of TP53 and MAPK pathway alterations. Less common ESR1 alterations falling outside the H11-H12 region seem to occur in tumors with higher TMB, deserving further investigation to understand their potential actionability.
Keywords: ESR1; breast cancer; genomic analysis; next-generation sequencing; precision medicine.
Copyright © 2024 The Author(s). Published by Elsevier Ltd.. All rights reserved.
Figures
References
-
- Burstein H.J. Systemic therapy for estrogen receptor–positive, HER2-negative breast cancer. N Engl J Med. 2020;383(26):2557–2570. - PubMed
-
- Yu F., Bender W. The mechanism of tamoxifen in breast cancer prevention. Breast Cancer Res. 2001;3(suppl 1):A74. - PubMed
-
- Emons G., Schally A.V. The use of luteinizing hormone releasing hormone agonists and antagonists in gynaecological cancers. Hum Reprod. 1994;9(7):1364–1379. - PubMed
-
- Miller W. Aromatase inhibitors: mechanism of action and role in the treatment of breast cancer. Semin Oncol. 2003;30:3–11. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

