Large-scale application of ClinGen-InSiGHT APC-specific ACMG/AMP variant classification criteria leads to substantial reduction in VUS
- PMID: 39357517
- PMCID: PMC11568752
- DOI: 10.1016/j.ajhg.2024.09.002
Large-scale application of ClinGen-InSiGHT APC-specific ACMG/AMP variant classification criteria leads to substantial reduction in VUS
Abstract
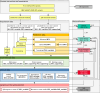
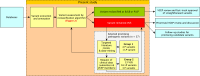
Pathogenic constitutional APC variants underlie familial adenomatous polyposis, the most common hereditary gastrointestinal polyposis syndrome. To improve variant classification and resolve the interpretative challenges of variants of uncertain significance (VUSs), APC-specific variant classification criteria were developed by the ClinGen-InSiGHT Hereditary Colorectal Cancer/Polyposis Variant Curation Expert Panel (VCEP) based on the criteria of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP). A streamlined algorithm using the APC-specific criteria was developed and applied to assess all APC variants in ClinVar and the International Society for Gastrointestinal Hereditary Tumours (InSiGHT) international reference APC Leiden Open Variation Database (LOVD) variant database, which included a total of 10,228 unique APC variants. Among the ClinVar and LOVD variants with an initial classification of (likely) benign or (likely) pathogenic, 94% and 96% remained in their original categories, respectively. In contrast, 41% ClinVar and 61% LOVD VUSs were reclassified into clinically meaningful classes, the vast majority as (likely) benign. The total number of VUSs was reduced by 37%. In 24 out of 37 (65%) promising APC variants that remained VUS despite evidence for pathogenicity, a data-mining-driven work-up allowed their reclassification as (likely) pathogenic. These results demonstrated that the application of APC-specific criteria substantially reduced the number of VUSs in ClinVar and LOVD. The study also demonstrated the feasibility of a systematic approach to variant classification in large datasets, which might serve as a generalizable model for other gene- or disease-specific variant interpretation initiatives. It also allowed for the prioritization of VUSs that will benefit from in-depth evidence collection. This subset of APC variants was approved by the VCEP and made publicly available through ClinVar and LOVD for widespread clinical use.
Keywords: ACMG/AMP variant classification guidelines; APC; Adenomatous polyposis coli; ClinGen; FAP; InSiGHT; familial adenomatous polyposis.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.E.P. is a member of the scientific advisory panel of Baylor Genetics Laboratories.
Figures




References
-
- Kinzler K.W., Nilbert M.C., Su L.K., Vogelstein B., Bryan T.M., Levy D.B., Smith K.J., Preisinger A.C., Hedge P., McKechnie D. Identification of FAP locus genes from chromosome 5q21. Science. 1991;253:661–665. - PubMed
-
- Groden J., Thliveris A., Samowitz W., Carlson M., Gelbert L., Albertsen H., Joslyn G., Stevens J., Spirio L., Robertson M. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. - PubMed
-
- WHO . In: Genetic Tumour Syndromes 5th ed. WCoTE B., editor. 2024. The Introduction to Genetic Tumour Syndromes.

