Molecular and functional landscape of malignant serous effusions for precision oncology
- PMID: 39358333
- PMCID: PMC11447229
- DOI: 10.1038/s41467-024-52694-8
Molecular and functional landscape of malignant serous effusions for precision oncology
Abstract
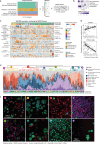
Personalized treatment for patients with advanced solid tumors critically depends on the deep characterization of tumor cells from patient biopsies. Here, we comprehensively characterize a pan-cancer cohort of 150 malignant serous effusion (MSE) samples at the cellular, molecular, and functional level. We find that MSE-derived cancer cells retain the genomic and transcriptomic profiles of their corresponding primary tumors, validating their use as a patient-relevant model system for solid tumor biology. Integrative analyses reveal that baseline gene expression patterns relate to global ex vivo drug sensitivity, while high-throughput drug-induced transcriptional changes in MSE samples are indicative of drug mode of action and acquired treatment resistance. A case study exemplifies the added value of multi-modal MSE profiling for patients who lack genetically stratified treatment options. In summary, our study provides a functional multi-omics view on a pan-cancer solid tumor cohort and underlines the feasibility and utility of MSE-based precision oncology.
© 2024. The Author(s).
Conflict of interest statement
B.S. was a scientific co-founder of Allcyte, which has been acquired by Exscientia. B.S. is a shareholder of Exscientia and a co-inventor on US patent application 15/514,045 relevant to the study. C.B. reports consulting or advisory role for AstraZeneca, Pfizer, Roche, Takeda, Janssen-Cilag, Boehringer-Ingelheim, Merck KGaA, Sanofi; research funding from Bayer; and travel, accommodation, and expenses from AstraZeneca, Takeda, Amgen. All of those are outside the presented work. H.M. is on advisory boards for Astra Zeneca, Stemline Therapeutics, Bayer, Amgen, Astella, MSD, Roche, and Merck. M.Z. receives research funds from Roche. P.P. is a scientific advisor for the company Biognosys AG (Zurich, Switzerland). All other authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

