ZIC2 and ZIC3 promote SWI/SNF recruitment to safeguard progression towards human primed pluripotency
- PMID: 39358345
- PMCID: PMC11447223
- DOI: 10.1038/s41467-024-52431-1
ZIC2 and ZIC3 promote SWI/SNF recruitment to safeguard progression towards human primed pluripotency
Abstract
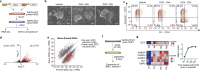
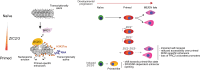
The primed epiblast acts as a transitional stage between the relatively homogeneous naïve epiblast and the gastrulating embryo. Its formation entails coordinated changes in regulatory circuits driven by transcription factors and epigenetic modifications. Using a multi-omic approach in human embryonic stem cell models across the spectrum of peri-implantation development, we demonstrate that the transcription factors ZIC2 and ZIC3 have overlapping but essential roles in opening primed-specific enhancers. Together, they are essential to facilitate progression to and maintain primed pluripotency. ZIC2/3 accomplish this by recruiting SWI/SNF to chromatin and loss of ZIC2/3 or degradation of SWI/SNF both prevent enhancer activation. Loss of ZIC2/3 also results in transcriptome changes consistent with perturbed Polycomb activity and a shift towards the expression of genes linked to differentiation towards the mesendoderm. Additionally, we find an intriguing dependency on the transcriptional machinery for sustained recruitment of ZIC2/3 over a subset of primed-hESC specific enhancers. Taken together, ZIC2 and ZIC3 regulate highly dynamic lineage-specific enhancers and collectively act as key regulators of human primed pluripotency.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- RGPIN-2018-04856/Gouvernement du Canada | Natural Sciences and Engineering Research Council of Canada (Conseil de Recherches en Sciences Naturelles et en Génie du Canada)
- PJT-166169/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
- PJT-173443/Gouvernement du Canada | Canadian Institutes of Health Research (Instituts de Recherche en Santé du Canada)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

