The role of TiO2 and gC3N4 bimetallic catalysts in boosting antibiotic resistance gene removal through photocatalyst assisted peroxone process
- PMID: 39358462
- PMCID: PMC11447026
- DOI: 10.1038/s41598-024-74147-4
The role of TiO2 and gC3N4 bimetallic catalysts in boosting antibiotic resistance gene removal through photocatalyst assisted peroxone process
Abstract
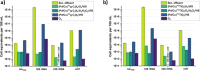
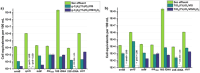
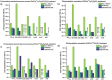
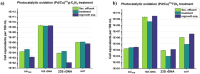
Antibiotics are extensively used in human medicine, aquaculture, and animal husbandry, leading to the release of antimicrobial resistance into the environment. This contributes to the rapid spread of antibiotic-resistant genes (ARGs), posing a significant threat to human health and aquatic ecosystems. Conventional wastewater treatment methods often fail to eliminate ARGs, prompting the adoption of advanced oxidation processes (AOPs) to address this growing risk. The study investigates the efficacy of visible light-driven photocatalytic systems utilizing two catalyst types (TiO2-Pd/Cu and g-C3N4-Pd/Cu), with a particular emphasis on their effectiveness in eliminating blaTEM, ermB, qnrS, tetM. intl1, 16 S rDNA and 23 S rDNA through photocatalytic ozonation and peroxone processes. Incorporating O3 into photocatalytic processes significantly enhances target removal efficiency, with the photocatalyst-assisted peroxone process emerging as the most effective AOP. The reemergence of targeted contaminants following treatment highlights the pivotal importance of AOPs and the meticulous selection of catalysts in ensuring sustained treatment efficacy. Furthermore, Polymerase Chain Reaction-Denaturing Gradient Gel Electrophoresis (PCR-DGGE) analysis reveals challenges in eradicating GC-rich bacteria with TiO2 and g-C3N4 processes, while slight differences in Cu/Pd loadings suggest g-C3N4-based ozonation improved antibacterial effectiveness. Terminal Restriction Fragment Length Polymorphism analysis highlights the efficacy of the photocatalyst-assisted peroxone process in treating diverse samples.
Keywords: Antibiotic-resistant bacteria; Antibiotic-resistant genes; Bimetallic catalysts; Photocatalyst-assisted peroxone process; Photocatalytic ozonation; Regrowth.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Zhang, M., Chen, S., Yu, X., Vikesland, P. & Pruden, A. Degradation of extracellular genomic, plasmid DNA and specific antibiotic resistance genes by chlorination. Front. Environ. Sci. Eng. 13 (2019).
-
- Bungau, S., Tit, D. M., Behl, T., Aleya, L. & Zaha, D. C. Aspects of excessive antibiotic consumption and environmental influences correlated with the occurrence of resistance to antimicrobial agents. Curr. Opin. Environ. Sci. Health. (2021). 10.1016/j.coesh.2020.10.012
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

