Tuft cells act as regenerative stem cells in the human intestine
- PMID: 39358509
- PMCID: PMC11499303
- DOI: 10.1038/s41586-024-07952-6
Tuft cells act as regenerative stem cells in the human intestine
Abstract
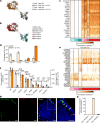
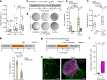
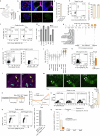
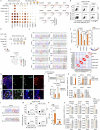
In mice, intestinal tuft cells have been described as a long-lived, postmitotic cell type. Two distinct subsets have been identified: tuft-1 and tuft-2 (ref. 1). By combining analysis of primary human intestinal resection material and intestinal organoids, we identify four distinct human tuft cell states, two of which overlap with their murine counterparts. We show that tuft cell development depends on the presence of Wnt ligands, and that tuft cell numbers rapidly increase on interleukin-4 (IL-4) and IL-13 exposure, as reported previously in mice2-4. This occurs through proliferation of pre-existing tuft cells, rather than through increased de novo generation from stem cells. Indeed, proliferative tuft cells occur in vivo both in fetal and in adult human intestine. Single mature proliferating tuft cells can form organoids that contain all intestinal epithelial cell types. Unlike stem and progenitor cells, human tuft cells survive irradiation damage and retain the ability to generate all other epithelial cell types. Accordingly, organoids engineered to lack tuft cells fail to recover from radiation-induced damage. Thus, tuft cells represent a damage-induced reserve intestinal stem cell pool in humans.
© 2024. The Author(s).
Conflict of interest statement
H.C. is the head of Pharma Research and Early Development at Roche, Basel and holds several patents related to organoids technology. His full disclosure is availble at
Figures






References
-
- Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature449, 1003–1007 (2007). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

