Connectomic reconstruction predicts visual features used for navigation
- PMID: 39358517
- PMCID: PMC11446847
- DOI: 10.1038/s41586-024-07967-z
Connectomic reconstruction predicts visual features used for navigation
Abstract
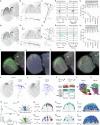
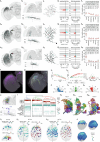
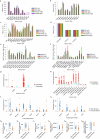
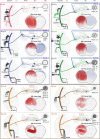
Many animals use visual information to navigate1-4, but how such information is encoded and integrated by the navigation system remains incompletely understood. In Drosophila melanogaster, EPG neurons in the central complex compute the heading direction5 by integrating visual input from ER neurons6-12, which are part of the anterior visual pathway (AVP)10,13-16. Here we densely reconstruct all neurons in the AVP using electron-microscopy data17. The AVP comprises four neuropils, sequentially linked by three major classes of neurons: MeTu neurons10,14,15, which connect the medulla in the optic lobe to the small unit of the anterior optic tubercle (AOTUsu) in the central brain; TuBu neurons9,16, which connect the AOTUsu to the bulb neuropil; and ER neurons6-12, which connect the bulb to the EPG neurons. On the basis of morphologies, connectivity between neural classes and the locations of synapses, we identify distinct information channels that originate from four types of MeTu neurons, and we further divide these into ten subtypes according to the presynaptic connections in the medulla and the postsynaptic connections in the AOTUsu. Using the connectivity of the entire AVP and the dendritic fields of the MeTu neurons in the optic lobes, we infer potential visual features and the visual area from which any ER neuron receives input. We confirm some of these predictions physiologically. These results provide a strong foundation for understanding how distinct sensory features can be extracted and transformed across multiple processing stages to construct higher-order cognitive representations.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures














Update of
-
Connectomic reconstruction predicts the functional organization of visual inputs to the navigation center of the Drosophila brain.bioRxiv [Preprint]. 2023 Nov 30:2023.11.29.569241. doi: 10.1101/2023.11.29.569241. bioRxiv. 2023. Update in: Nature. 2024 Oct;634(8032):181-190. doi: 10.1038/s41586-024-07967-z. PMID: 38076786 Free PMC article. Updated. Preprint.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

