Neural circuit mechanisms underlying context-specific halting in Drosophila
- PMID: 39358520
- PMCID: PMC11446846
- DOI: 10.1038/s41586-024-07854-7
Neural circuit mechanisms underlying context-specific halting in Drosophila
Abstract
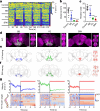
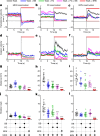
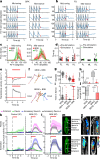
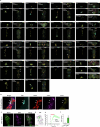
Walking is a complex motor programme involving coordinated and distributed activity across the brain and the spinal cord. Halting appropriately at the correct time is a critical component of walking control. Despite progress in identifying neurons driving halting1-6, the underlying neural circuit mechanisms responsible for overruling the competing walking state remain unclear. Here, using connectome-informed models7-9 and functional studies, we explain two fundamental mechanisms by which Drosophila implement context-appropriate halting. The first mechanism ('walk-OFF') relies on GABAergic neurons that inhibit specific descending walking commands in the brain, whereas the second mechanism ('brake') relies on excitatory cholinergic neurons in the nerve cord that lead to an active arrest of stepping movements. We show that two neurons that deploy the walk-OFF mechanism inhibit distinct populations of walking-promotion neurons, leading to differential halting of forward walking or turning. The brake neurons, by constrast, override all walking commands by simultaneously inhibiting descending walking-promotion neurons and increasing the resistance at the leg joints. We characterized two behavioural contexts in which the distinct halting mechanisms were used by the animal in a mutually exclusive manner: the walk-OFF mechanism was engaged for halting during feeding and the brake mechanism was engaged for halting and stability during grooming.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures










References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

