Whole-brain annotation and multi-connectome cell typing of Drosophila
- PMID: 39358521
- PMCID: PMC11446831
- DOI: 10.1038/s41586-024-07686-5
Whole-brain annotation and multi-connectome cell typing of Drosophila
Abstract
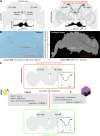
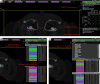
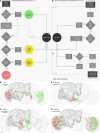
The fruit fly Drosophila melanogaster has emerged as a key model organism in neuroscience, in large part due to the concentration of collaboratively generated molecular, genetic and digital resources available for it. Here we complement the approximately 140,000 neuron FlyWire whole-brain connectome1 with a systematic and hierarchical annotation of neuronal classes, cell types and developmental units (hemilineages). Of 8,453 annotated cell types, 3,643 were previously proposed in the partial hemibrain connectome2, and 4,581 are new types, mostly from brain regions outside the hemibrain subvolume. Although nearly all hemibrain neurons could be matched morphologically in FlyWire, about one-third of cell types proposed for the hemibrain could not be reliably reidentified. We therefore propose a new definition of cell type as groups of cells that are each quantitatively more similar to cells in a different brain than to any other cell in the same brain, and we validate this definition through joint analysis of FlyWire and hemibrain connectomes. Further analysis defined simple heuristics for the reliability of connections between brains, revealed broad stereotypy and occasional variability in neuron count and connectivity, and provided evidence for functional homeostasis in the mushroom body through adjustments of the absolute amount of excitatory input while maintaining the excitation/inhibition ratio. Our work defines a consensus cell type atlas for the fly brain and provides both an intellectual framework and open-source toolchain for brain-scale comparative connectomics.
© 2024. The Author(s).
Conflict of interest statement
H.S.S. declares a financial interest in Zetta AI. The other authors declare no competing interests.
Figures















Update of
-
Whole-brain annotation and multi-connectome cell typing quantifies circuit stereotypy in Drosophila.bioRxiv [Preprint]. 2023 Jul 15:2023.06.27.546055. doi: 10.1101/2023.06.27.546055. bioRxiv. 2023. Update in: Nature. 2024 Oct;634(8032):139-152. doi: 10.1038/s41586-024-07686-5. PMID: 37425808 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

