MHC Hammer reveals genetic and non-genetic HLA disruption in cancer evolution
- PMID: 39358601
- PMCID: PMC11525181
- DOI: 10.1038/s41588-024-01883-8
MHC Hammer reveals genetic and non-genetic HLA disruption in cancer evolution
Abstract
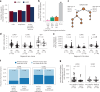
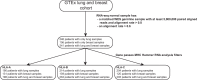
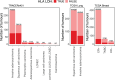
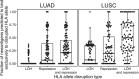
Disruption of the class I human leukocyte antigen (HLA) molecules has important implications for immune evasion and tumor evolution. We developed major histocompatibility complex loss of heterozygosity (LOH), allele-specific mutation and measurement of expression and repression (MHC Hammer). We identified extensive variability in HLA allelic expression and pervasive HLA alternative splicing in normal lung and breast tissue. In lung TRACERx and lung and breast TCGA cohorts, 61% of lung adenocarcinoma (LUAD), 76% of lung squamous cell carcinoma (LUSC) and 35% of estrogen receptor-positive (ER+) cancers harbored class I HLA transcriptional repression, while HLA tumor-enriched alternative splicing occurred in 31%, 11% and 15% of LUAD, LUSC and ER+ cancers. Consistent with the importance of HLA dysfunction in tumor evolution, in LUADs, HLA LOH was associated with metastasis and LUAD primary tumor regions seeding a metastasis had a lower effective neoantigen burden than non-seeding regions. These data highlight the extent and importance of HLA transcriptomic disruption, including repression and alternative splicing in cancer evolution.
© 2024. The Author(s).
Conflict of interest statement
C.P. holds a patent pending in determining HLA disruption (PCT/EP2023/059039). K.K.D. provided consultancy services to Achilles Therapeutics UK. N.K. receives research support from AstraZeneca. K.L. has a patent on InDel burden and CPI response pending and speaker fees from Roche tissue diagnostics, research funding from CRUK TDL–Ono–LifeArc alliance, Genesis Therapeutics and consulting roles with Ellipses Pharma, Monopteros and Kynos Therapeutics. D.A.M. reports speaker fees from Eli Lilly, AstraZeneca and Takeda Pharmaceuticals; consultancy fees from AstraZeneca, Thermo Fisher Scientific, Takeda Pharmaceuticals, Amgen, Janssen, MIM Software, Bristol Myers Squibb and Eli Lilly; and has received educational support from Takeda Pharmaceuticals and Amgen. M.J.-H. has received funding from CRUK, NIH National Cancer Institute, IASLC International Lung Cancer Foundation, Lung Cancer Research Foundation, Rosetrees Trust, UKI NETs and NIHR. M.J-H. has consulted for, and is a member of, the Achilles Therapeutics Scientific Advisory Board and Steering Committee and has received speaker honoraria from Pfizer, Astex Pharmaceuticals, Oslo Cancer Cluster, Bristol Myers Squibb and Genentech. M.J.-.H. is listed as a co-inventor on a European patent application relating to methods to detect lung cancer PCT/US2017/028013). This patent has been licensed to commercial entities, and under terms of employment, M.J.-H. is due a share of any revenue generated from such license(s) and is also listed as a co-inventor on the GB priority patent application (GB2400424.4) with title—Treatment and Prevention of Lung Cancer. C.S. acknowledges grants from AstraZeneca, Boehringer-Ingelheim, Bristol Myers Squibb, Pfizer, Roche-Ventana, Invitae (previously Archer Dx—collaboration in minimal residual disease sequencing technologies), Ono Pharmaceutical and Personalis. He is the chief investigator for the AZ MeRmaiD 1 and 2 clinical trials and is the Steering Committee Chair. He is also the cochief investigator of the NHS Galleri trial funded by GRAIL and a paid member of GRAIL’s Scientific Advisory Board. He receives consultant fees from Achilles Therapeutics (also a SAB member), Bicycle Therapeutics (also a SAB member), Genentech, Medicxi, China Innovation Centre of Roche (CICoR) formerly Roche Innovation Centre—Shanghai, Metabomed (until July 2022), Relay Therapeutics SAB member, Saga Diagnostics SAB member and the Sarah Cannon Research Institute. He has received honoraria from Amgen, AstraZeneca, Bristol Myers Squibb, GlaxoSmithKline, Illumina, MSD, Novartis, Pfizer and Roche-Ventana. He has previously held stock options in Apogen Biotechnologies and GRAIL; currently has stock options in Epic Bioscience, Bicycle Therapeutics, and Relay Therapeutics; and has stock options and is cofounder of Achilles Therapeutics. He declares a patent application for methods to lung cancer (PCT/US2017/028013); targeting neoantigens (PCT/EP2016/059401); identifying patent response to immune checkpoint blockade (PCT/EP2016/071471); methods for lung cancer detection (US20190106751A1); identifying patients who respond to cancer treatment (PCT/GB2018/051912); determining HLA LOH (PCT/GB2018/052004); predicting survival rates of patients with cancer (PCT/GB2020/050221); and methods and systems for tumor monitoring (PCT/EP2022/077987). He is an inventor of a European patent application (PCT/GB2017/053289) relating to assay technology to detect tumor recurrence. This patent has been licensed to a commercial entity, and under their terms of employment, he is due a revenue share of any revenue generated from such license(s). N.M. has stock options in and has consulted for Achilles Therapeutics and holds a European patent in determining HLA LOH (PCT/GB2018/052004), a patent pending in determining HLA disruption (PCT/EP2023/059039), and is a co-inventor to a patent to identify responders to cancer treatment (PCT/GB2018/051912). The remaining authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

