A new class of capsid-targeting inhibitors that specifically block HIV-1 nuclear import
- PMID: 39358603
- PMCID: PMC11555092
- DOI: 10.1038/s44321-024-00143-w
A new class of capsid-targeting inhibitors that specifically block HIV-1 nuclear import
Abstract
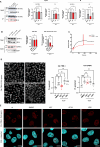
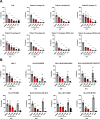
HIV-1 capsids cross nuclear pore complexes (NPCs) by engaging with the nuclear import machinery. To identify compounds that inhibit HIV-1 nuclear import, we screened drugs in silico on a three-dimensional model of a CA hexamer bound by Transportin-1 (TRN-1). Among hits, compound H27 inhibited HIV-1 with a low micromolar IC50. Unlike other CA-targeting compounds, H27 did not alter CA assembly or disassembly, inhibited nuclear import specifically, and retained antiviral activity against PF74- and Lenacapavir-resistant mutants. The differential sensitivity of divergent primate lentiviral capsids, capsid stability and H27 escape mutants, together with structural analyses, suggest that H27 makes multiple low affinity contacts with assembled capsid. Interaction experiments indicate that H27 may act by preventing CA from engaging with components of the NPC machinery such as TRN-1. H27 exhibited good metabolic stability in vivo and was efficient against different subtypes and circulating recombinant forms from treatment-naïve patients as well as strains resistant to the four main classes of antiretroviral drugs. This work identifies compounds that demonstrate a novel mechanism of action by specifically blocking HIV-1 nuclear import.
Keywords: Antiretroviral Therapy; Capsid; Drug Discovery; HIV-1; Nuclear Import.
© 2024. The Author(s).
Conflict of interest statement
Figures









References
-
- Bhargava A, Lahaye X, Manel N (2018) Let me in: control of HIV nuclear entry at the nuclear envelope. Cytokine Growth Factor Rev 40:59–67 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

