Pre-clinical development of AP4B1 gene replacement therapy for hereditary spastic paraplegia type 47
- PMID: 39358605
- PMCID: PMC11554807
- DOI: 10.1038/s44321-024-00148-5
Pre-clinical development of AP4B1 gene replacement therapy for hereditary spastic paraplegia type 47
Abstract
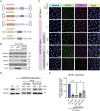
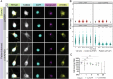
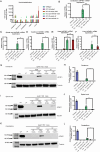
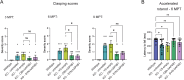
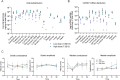
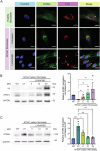
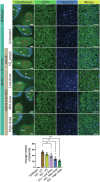
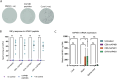
Spastic paraplegia 47 (SPG47) is a neurological disorder caused by mutations in the adaptor protein complex 4 β1 subunit (AP4B1) gene leading to AP-4 complex deficiency. SPG47 is characterised by progressive spastic paraplegia, global developmental delay, intellectual disability and epilepsy. Gene therapy aimed at restoring functional AP4B1 protein levels is a rational therapeutic strategy to ameliorate the disease phenotype. Here we report that a single delivery of adeno-associated virus serotype 9 expressing hAP4B1 (AAV9/hAP4B1) into the cisterna magna leads to widespread gene transfer and restoration of various hallmarks of disease, including AP-4 cargo (ATG9A) mislocalisation, calbindin-positive spheroids in the deep cerebellar nuclei, anatomical brain defects and motor dysfunction, in an SPG47 mouse model. Furthermore, AAV9/hAP4B1-based gene therapy demonstrated a restoration of plasma neurofilament light (NfL) levels of treated mice. Encouraged by these preclinical proof-of-concept data, we conducted IND-enabling studies, including immunogenicity and GLP non-human primate (NHP) toxicology studies. Importantly, NHP safety and biodistribution study revealed no significant adverse events associated with the therapeutic intervention. These findings provide evidence of both therapeutic efficacy and safety, establishing a robust basis for the pursuit of an IND application for clinical trials targeting SPG47 patients.
Keywords: AAV; AP4B1; Gene Therapy; HSP; SPG47.
© 2024. The Author(s).
Conflict of interest statement
Figures
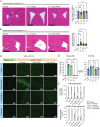
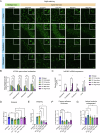
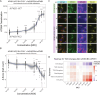
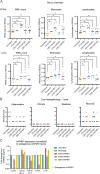


References
-
- Abou Jamra R, Philippe O, Raas-Rothschild A, Eck SH, Graf E, Buchert R, Borck G, Ekici A, Brockschmidt FF, Nöthen MM, Munnich A, Strom TM, Reis A, Colleaux L (2011) Adaptor protein complex 4 deficiency causes severe autosomal-recessive intellectual disability, progressive spastic paraplegia, shy character, and short stature. Am J Hum Genet 88(6):788–795 - PMC - PubMed
-
- Alecu JE, Saffari A, Ziegler M, Jordan C, Tam A, Kim S, Leung E, Szczaluba K, Mierzewska H, King SD, Santorelli FM, Yoon G, Trombetta B, Kivisäkk P, Zhang B, Sahin M, Ebrahimi-Fakhari D (2023) Plasma neurofilament light chain is elevated in adaptor protein complex 4-related hereditary spastic paraplegia. Mov Disord 38(9):1742–1750 - PMC - PubMed
-
- Bauer P, Leshinsky-Silver E, Blumkin L, Schlipf N, Schröder C, Schicks J, Lev D, Riess O, Lerman-Sagie T, Schöls L (2012) Mutation in the AP4B1 gene cause hereditary spastic paraplegia type 47 (SPG47). Neurogenetics 13(1):73–76 - PubMed
-
- Behne R, Teinert J, Wimmer M, D’Amore A, Davies AK, Scarrott JM, Eberhardt K, Brechmann B, Chen IP, Buttermore ED et al (2020) Adaptor protein complex 4 deficiency: a paradigm of childhood-onset hereditary spastic paraplegia caused by defective protein trafficking. Hum Mol Genet 29(2):320–334 - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

