Genus-targeted markers for the taxonomic identification and monitoring of coagulase-positive and coagulase-negative Staphylococcus species
- PMID: 39358646
- PMCID: PMC11447098
- DOI: 10.1007/s11274-024-04121-9
Genus-targeted markers for the taxonomic identification and monitoring of coagulase-positive and coagulase-negative Staphylococcus species
Abstract
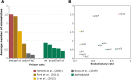
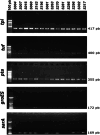
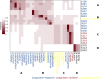
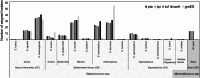
The Staphylococcus genus comprises multiple pathogenic and opportunistic species that represent a risk to public health. Epidemiological studies require accurate taxonomic classification of isolates with enough resolution to distinguish clonal complexes. Unfortunately, 16 S rRNA molecular analysis and phenotypic characterization cannot distinguish all species and do not offer enough resolution to assess intraspecific diversity. Other approaches, such as Multilocus Sequence Tagging, provide higher resolution; however, they have been developed for Staphylococcus aureus and a few other species. Here, we developed a set of genus-targeted primers using five orthologous genes (pta, tuf, tpi, groEs, and sarA) to identify all Staphylococcus species within the genus. The primers were initially evaluated using 20 strains from the Collection of Microorganisms of Interest in Animal Health from AGROSAVIA (CMISA), and their amplified sequences were compared to a set of 33 Staphylococcus species. This allowed the taxonomic identification of the strains even on close species and the establishment of intraspecies diversity. To enhance the scope and cost-effectiveness of the proposed strategy, we customized the primer sets for an Illumina paired-end amplicon protocol, enabling gene multiplexing. We assessed five genes across 177 strains, generating 880 paired-end libraries from the CMISA. This approach significantly reduced sequencing costs, as all libraries can be efficiently sequenced in a single MiSeq run at a fraction (one-fourth or less) of the cost associated with Sanger sequencing. In summary, this method can be used for precise identification and diversity analysis of Staphylococcus species, offering an advancement over traditional techniques in both resolution and cost-effectiveness.
Keywords: Epidemiology; Molecular typing; Phylogenetics; Protein-coding genes; Staphylococcus.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Abdul-Aziz A, Mohamad SAS, Abdullah MFF (2015) Identification of coagulase-negative staphylococci by SodA gene sequence analysis. Curr Res Bacteriol 8:48–61. 10.3923/crb.2015.48.61
-
- Adkins PRF, Middleton JR, Calcutt MJ et al (2017) Species identification and strain typing of Staphylococcus agnetis and Staphylococcus hyicus isolates from bovine milk by use of a novel multiplex PCR assay and pulsed-field gel electrophoresis. J Clin Microbiol 55:1778–1788. 10.1128/JCM.02239-16 - PMC - PubMed
-
- Altschul SF, Gish W, Miller W et al (1990) Basic Local Alignment search toll. J Mol Biol 215:403–410 - PubMed
-
- Asante J, Amoako DG, Abia ALK et al (2020) Review of clinically and epidemiologically relevant Coagulase-negative Staphylococci in Africa. Microb Drug Resist 00. 10.1089/mdr.2019.0381 - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

