A single amplified genome catalog reveals the dynamics of mobilome and resistome in the human microbiome
- PMID: 39358771
- PMCID: PMC11446047
- DOI: 10.1186/s40168-024-01903-z
A single amplified genome catalog reveals the dynamics of mobilome and resistome in the human microbiome
Abstract
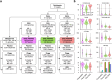
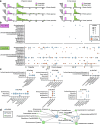
Background: The increase in metagenome-assembled genomes (MAGs) has advanced our understanding of the functional characterization and taxonomic assignment within the human microbiome. However, MAGs, as population consensus genomes, often aggregate heterogeneity among species and strains, thereby obfuscating the precise relationships between microbial hosts and mobile genetic elements (MGEs). In contrast, single amplified genomes (SAGs) derived via single-cell genome sequencing can capture individual genomic content, including MGEs.
Results: We introduce the first substantial SAG dataset (bbsag20) from the human oral and gut microbiome, comprising 17,202 SAGs above medium-quality without co-assembly. This collection unveils a diversity of bacterial lineages across 312 oral and 647 gut species, demonstrating different taxonomic compositions from MAGs. Moreover, the SAGs showed cellular-level evidence of the translocation of oral bacteria to the gut. We also identified broad-host-range MGEs harboring antibiotic resistance genes (ARGs), which were not detected in the MAGs.
Conclusions: The difference in taxonomic composition between SAGs and MAGs indicates that combining both methods would be effective in expanding the genome catalog. By connecting mobilomes and resistomes in individual samples, SAGs could meticulously chart a dynamic network of ARGs on MGEs, pinpointing potential ARG reservoirs and their spreading patterns in the microbial community. Video Abstract.
© 2024. The Author(s).
Conflict of interest statement
MH is a founder and shareholder in bitBiome, Inc., which provides single-cell genomics services using the SAG-gel workflow as bit-MAP. TKS, KA, TS, TE, KK, and AM are employed at bitBiome, Inc. MH, TS, TE, KK, and KA are inventors on patent applications submitted by bitBiome, Inc., covering the technique for single-cell sequencing.
Figures

Similar articles
-
Metagenomic approach revealed the mobility and co-occurrence of antibiotic resistomes between non-intensive aquaculture environment and human.Microbiome. 2024 Jun 14;12(1):107. doi: 10.1186/s40168-024-01824-x. Microbiome. 2024. PMID: 38877573 Free PMC article.
-
Antimicrobial resistance and its risks evaluation in wetlands on the Qinghai-Tibetan Plateau.Ecotoxicol Environ Saf. 2024 Sep 1;282:116699. doi: 10.1016/j.ecoenv.2024.116699. Epub 2024 Jul 8. Ecotoxicol Environ Saf. 2024. PMID: 38981389
-
Antibiotic resistance gene dynamics in the commensal infant gut microbiome over the first year of life.Sci Rep. 2024 Aug 12;14(1):18701. doi: 10.1038/s41598-024-69275-w. Sci Rep. 2024. PMID: 39134593 Free PMC article.
-
Genome-resolved metagenomics: a game changer for microbiome medicine.Exp Mol Med. 2024 Jul;56(7):1501-1512. doi: 10.1038/s12276-024-01262-7. Epub 2024 Jul 1. Exp Mol Med. 2024. PMID: 38945961 Free PMC article. Review.
-
Probing the Mobilome: Discoveries in the Dynamic Microbiome.Trends Microbiol. 2021 Feb;29(2):158-170. doi: 10.1016/j.tim.2020.05.003. Epub 2020 May 11. Trends Microbiol. 2021. PMID: 32448763 Review.
Cited by
-
The Neonatal Microbiome: Implications for Amyotrophic Lateral Sclerosis and Other Neurodegenerations.Brain Sci. 2025 Feb 14;15(2):195. doi: 10.3390/brainsci15020195. Brain Sci. 2025. PMID: 40002527 Free PMC article. Review.
References
-
- Lloyd-Price J, Arze C, Ananthakrishnan AN, Schirmer M, Avila-Pacheco J, Poon TW, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–62 https://www.nature.com/articles/s41586-019-1237-9. - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

