ATF3-mediated transactivation of CXCL14 in HSCs during liver fibrosis
- PMID: 39358917
- PMCID: PMC11446984
- DOI: 10.1002/ctm2.70040
ATF3-mediated transactivation of CXCL14 in HSCs during liver fibrosis
Abstract
Background and aims: Myofibroblasts, the primary producers of extracellular matrix, primarily originate from hepatic stellate cells (HSCs), and their activation plays a pivotal role in liver fibrosis. This study aimed to investigate the function of CXC motif ligand 14 (CXCL14) in the progression of liver fibrosis.
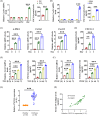
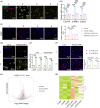
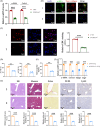
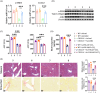
Approach and results: CXCL14 knockdown significantly reduced the extent of liver fibrosis. Using Ingenuity pathway analysis and qRT-PCR, activating transcription factor 3 (ATF3) was identified as a key upstream regulator of CXCL14 expression. Mechanistically, ATF3 was shown to bind to the CXCL14 promoter, promoting its transactivation by TGF-β in HSCs. Notably, both global CXCL14 deletion (CXCL14-/-) and HSC/myofibroblast-specific CXCL14 knockdown significantly attenuated liver fibrosis in mice. RNA-seq comparisons between CXCL14-/- and WT mice highlighted Jak2 as the most significantly downregulated gene, implicating its role in the antifibrotic effects of CXCL14 suppression on HSC inactivation. Moreover, Jak2 overexpression reversed the inhibition of liver fibrosis caused by CXCL14 knockdown in vivo.
Conclusions: These findings unveil a novel ATF3/CXCL14/Jak2 signalling axis in liver fibrosis, presenting potential therapeutic targets for the disease.
Keywords: CXCL14; hepatic stellate cell; liver fibrosis; transcriptional regulation.
© 2024 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare no financial conflicts of interest or personal relationships that could have influenced the findings presented in this paper.
Figures




References
-
- Kisseleva T, Brenner D. Molecular and cellular mechanisms of liver fibrosis and its regression. Nat Rev Gastroenterol Hepatol. 2021;18(3):151‐166. - PubMed
-
- She H, Xiong S, Hazra S, Tsukamoto H. Adipogenic transcriptional regulation of hepatic stellate cells. J Biol Chem. 2005;280(6):4959‐4967. - PubMed
-
- Qiang G, Zhang L, Yang X, et al. Effect of valsartan on the pathological progression of hepatic fibrosis in rats with type 2 diabetes. Eur J Pharmacol. 2012;685(1‐3):156‐164. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
