Inflammation-induced loss of CFTR-expressing airway ionocytes in non-eosinophilic asthma
- PMID: 39358991
- PMCID: PMC11688627
- DOI: 10.1111/resp.14833
Inflammation-induced loss of CFTR-expressing airway ionocytes in non-eosinophilic asthma
Abstract
Background and objective: Severe asthma is a heterogeneous disease with subtype classification according to dominant airway infiltrates, including eosinophilic (Type 2 high), or non-eosinophilic asthma. Non-eosinophilic asthma is further divided into paucigranulocytic or neutrophilic asthma characterized by elevated neutrophils, and mixed Type 1 and Type 17 cytokines in the airways. Severe non-eosinophilic asthma has few effective treatments and many patients do not qualify for biologic therapies. The cystic fibrosis transmembrane conductance regulator (CFTR) is dysregulated in multiple respiratory diseases including cystic fibrosis and chronic obstructive pulmonary disease and has proven a valuable therapeutic target. We hypothesized that the CFTR may also play a role in non-eosinophilic asthma.
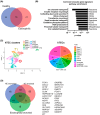
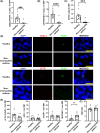
Methods: Patient-derived human bronchial epithelial cells (hBECs) were isolated and differentiated at the air-liquid interface. Single cell RNA-sequencing (scRNAseq) was used to identify epithelial cell subtypes and transcriptional activity. Ion transport was investigated with Ussing chambers and immunofluorescent quantification of ionocyte abundance in human airway epithelial cells and murine models of asthma.
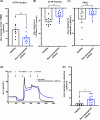
Results: We identified that hBECs from patients with non-eosinophilic asthma had reduced CFTR function, and did not differentiate into CFTR-expressing ionocytes compared to those from eosinophilic asthma or healthy donors. Similarly, ionocytes were also diminished in the airways of a murine model of neutrophilic-dominant but not eosinophilic asthma. Treatment of hBECs from healthy donors with a neutrophilic asthma-like inflammatory cytokine mixture led to a reduction in ionocytes.
Conclusion: Inflammation-induced loss of CFTR-expressing ionocytes in airway cells from non-eosinophilic asthma may represent a key feature of disease pathogenesis and a novel drug target.
Keywords: CFTR; airway epithelium; ionocytes; mucous; neutrophils; severe asthma; single cell RNA‐sequencing.
© 2024 The Author(s). Respirology published by John Wiley & Sons Australia, Ltd on behalf of Asian Pacific Society of Respirology.
Conflict of interest statement
G. E. K. and L. C. have performed university contract research for Krystal Biotech on CFTR in cystic fibrosis but not related to this work, and not involved in the funding of this work. P. A. B. W. has received honoraria from GSK, Astra Zeneca, Sanofi, Novartis and Vertex but not for this work. P. A. B. W. is a committee member of the National Asthma Council of Australia. The other authors declare no conflicts of interest.
Figures



References
-
- Brusselle GG, Koppelman GH. Biologic therapies for severe asthma. N Engl J Med. 2022;386(2):157–171. - PubMed
-
- Baines KJ, Simpson JL, Wood LG, Scott RJ, Fibbens NL, Powell H, et al. Sputum gene expression signature of 6 biomarkers discriminates asthma inflammatory phenotypes. J Allergy Clin Immunol. 2014;133(4):997–1007. - PubMed
-
- Barnes PJ. Inflammatory mechanisms in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2016;138(1):16–27. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

