Potential applications of microbial genomics in nuclear non-proliferation
- PMID: 39360321
- PMCID: PMC11445143
- DOI: 10.3389/fmicb.2024.1410820
Potential applications of microbial genomics in nuclear non-proliferation
Abstract
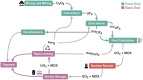
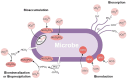
As nuclear technology evolves in response to increased demand for diversification and decarbonization of the energy sector, new and innovative approaches are needed to effectively identify and deter the proliferation of nuclear arms, while ensuring safe development of global nuclear energy resources. Preventing the use of nuclear material and technology for unsanctioned development of nuclear weapons has been a long-standing challenge for the International Atomic Energy Agency and signatories of the Treaty on the Non-Proliferation of Nuclear Weapons. Environmental swipe sampling has proven to be an effective technique for characterizing clandestine proliferation activities within and around known locations of nuclear facilities and sites. However, limited tools and techniques exist for detecting nuclear proliferation in unknown locations beyond the boundaries of declared nuclear fuel cycle facilities, representing a critical gap in non-proliferation safeguards. Microbiomes, defined as "characteristic communities of microorganisms" found in specific habitats with distinct physical and chemical properties, can provide valuable information about the conditions and activities occurring in the surrounding environment. Microorganisms are known to inhabit radionuclide-contaminated sites, spent nuclear fuel storage pools, and cooling systems of water-cooled nuclear reactors, where they can cause radionuclide migration and corrosion of critical structures. Microbial transformation of radionuclides is a well-established process that has been documented in numerous field and laboratory studies. These studies helped to identify key bacterial taxa and microbially-mediated processes that directly and indirectly control the transformation, mobility, and fate of radionuclides in the environment. Expanding on this work, other studies have used microbial genomics integrated with machine learning models to successfully monitor and predict the occurrence of heavy metals, radionuclides, and other process wastes in the environment, indicating the potential role of nuclear activities in shaping microbial community structure and function. Results of this previous body of work suggest fundamental geochemical-microbial interactions occurring at nuclear fuel cycle facilities could give rise to microbiomes that are characteristic of nuclear activities. These microbiomes could provide valuable information for monitoring nuclear fuel cycle facilities, planning environmental sampling campaigns, and developing biosensor technology for the detection of undisclosed fuel cycle activities and proliferation concerns.
Keywords: environmental bioindicators; microbial ecology; microbial genomics; nuclear non-proliferation; radionuclides; systems biology.
Copyright © 2024 MacGregor, Fukai, Ash, Arkin and Hazen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
-
- Anderson R. T., Vrionis H. A., Ortiz-Bernad I., Resch C. T., Long P. E., Dayvault R., et al. . (2003). Stimulating the In Situ Activity of Geobacter Species to Remove Uranium from the Groundwater of a Uranium-Contaminated Aquifer. Appl. Environ. Microbiol. 69, 5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003 - DOI - PMC - PubMed
-
- Badora A., Kud K., Woźniak M. (2021). Nuclear energy perception and ecological attitudes. Energies 14:322. doi: 10.3390/en14144322 - DOI
-
- Bagwell C. E., Bhat S., Hawkins G. M., Smith B. W., Biswas T., Hoover T. R., et al. . (2008). Survival in Nuclear Waste, Extreme Resistance, and Potential Applications Gleaned from the Genome Sequence of Kineococcus radiotolerans SRS30216. PLoS One 3:878. doi: 10.1371/journal.pone.0003878 - DOI - PMC - PubMed
-
- Barton F., Shaw S., Morris K., Graham J., Lloyd J. R. (2022). Impact and control of fouling in radioactive environments. Prog. Nucl. Energy 148:104215. doi: 10.1016/j.pnucene.2022.104215 - DOI
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous