Impact of weight variation on the microbiome of yak dams and calves
- PMID: 39360324
- PMCID: PMC11446105
- DOI: 10.3389/fmicb.2024.1465992
Impact of weight variation on the microbiome of yak dams and calves
Abstract
Introduction: Limited information exists regarding the microbiome composition of yak calves of varying weights. Therefore, this study aimed to investigate the microbiomes of mother-calf pairs with different weight profiles.
Methods: Fecal and blood samples were collected from both lower-weight (CB) and higher-weight (HB) yak calves, along with their corresponding female yaks (CA, HA).
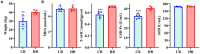
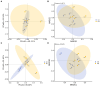
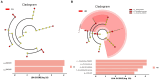
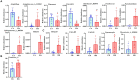
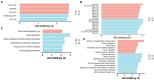
Results: The results revealed significantly higher levels of T-AOC (total antioxidant capacity) and GSH-Px (glutathione peroxidase) in HB animals (p < 0.001). Sequencing yielded 652,181 and 643,369 filtered reads in female and calf yaks, respectively. Alpha diversity analysis indicated that Chao1, Faith_pd, and Observed species were significantly higher in CA compared to HA (p < 0.01). Furthermore, nine genera were notably different between HA and CA yaks, including Avispirillum, Fimenecus, CAG-1031, Odoribacter 865974, and Jeotgalicoccus A 310962. Compared to CB yaks, CA animals exhibited significant differences in one phylum and six genera, including CAG-485 (p < 0.05), CAG-83 (p < 0.01), Copromorpha (p < 0.01), Phocaeicola A 858004 (p < 0.05), and UBA2253 (p < 0.05).
Conclusion: In summary, higher-weight yak calves demonstrated increased oxidative resistance, and weight profiles were linked to the microbiomes of both female yaks and their calves. These findings offer valuable insights for optimizing yak breeding practices in high-altitude regions.
Keywords: calf; microbiota; oxidative resistance; weight; yak.
Copyright © 2024 Wang, Basang, Pingcuo, Jiang, Sun, Nawaz, Cidan, Liu, Zhu and Luosang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Al Bataineh M. T., Künstner A., Dash N. R., Alsafar H. S., Ragab M., Schmelter F., et al. . (2023). Uncovering the relationship between gut microbial dysbiosis, metabolomics, and dietary intake in type 2 diabetes mellitus and in healthy volunteers: a multi-omics analysis. Sci. Rep. 13:45066. doi: 10.1038/s41598-023-45066-7 - DOI - PMC - PubMed
-
- Bogaert D., van Beveren G. J., de Koff E. M., Lusarreta Parga P., Balcazar Lopez C. E., Koppensteiner L., et al. . (2023). Mother-to-infant microbiota transmission and infant microbiota development across multiple body sites. Cell Host Microbe 31, 447–460.e6. doi: 10.1016/j.chom.2023.01.018, PMID: - DOI - PubMed
-
- Bowerman K. L., Varelias A., Lachner N., Kuns R. D., Hill G. R., Hugenholtz P. (2020). Continuous pre-and post-transplant exposure to a disease-associated gut microbiome promotes hyper-acute graft-versus-host disease in wild-type mice. Gut Microbes 11, 754–770. doi: 10.1080/19490976.2019.1705729, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

