Modulation of UPF1 catalytic activity upon interaction of SARS-CoV-2 Nucleocapsid protein with factors involved in nonsense mediated-mRNA decay
- PMID: 39360627
- PMCID: PMC11602160
- DOI: 10.1093/nar/gkae829
Modulation of UPF1 catalytic activity upon interaction of SARS-CoV-2 Nucleocapsid protein with factors involved in nonsense mediated-mRNA decay
Abstract
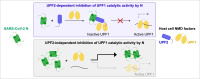
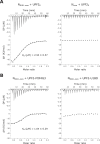
The RNA genome of the SARS-CoV-2 virus encodes for four structural proteins, 16 non-structural proteins and nine putative accessory factors. A high throughput analysis of interactions between human and SARS-CoV-2 proteins identified multiple interactions of the structural Nucleocapsid (N) protein with RNA processing factors. The N-protein, which is responsible for packaging of the viral genomic RNA was found to interact with two RNA helicases, UPF1 and MOV10 that are involved in nonsense-mediated mRNA decay (NMD). Using a combination of biochemical and biophysical methods, we investigated the interaction of the SARS-CoV-2 N-protein with NMD factors at a molecular level. Our studies led us to identify the core NMD factor, UPF2, as an interactor of N. The viral N-protein engages UPF2 in multipartite interactions and can negate the stimulatory effect of UPF2 on UPF1 catalytic activity. N also inhibits UPF1 ATPase and unwinding activities by competing in binding to the RNA substrate. We further investigate the functional implications of inhibition of UPF1 catalytic activity by N in mammalian cells. The interplay of SARS-CoV-2 N with human UPF1 and UPF2 does not affect decay of host cell NMD targets but might play a role in stabilizing the viral RNA genome.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

