Altered dNTP pools accelerate tumor formation in mice
- PMID: 39360631
- PMCID: PMC11551754
- DOI: 10.1093/nar/gkae843
Altered dNTP pools accelerate tumor formation in mice
Abstract
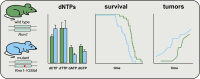
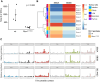
Alterations in deoxyribonucleoside triphosphate (dNTP) pools have been linked to increased mutation rates and genome instability in unicellular organisms and cell cultures. However, the role of dNTP pool changes in tumor development in mammals remains unclear. In this study, we present a mouse model with a point mutation at the allosteric specificity site of ribonucleotide reductase, RRM1-Y285A. This mutation reduced ribonucleotide reductase activity, impairing the synthesis of deoxyadenosine triphosphate (dATP) and deoxyguanosine triphosphate (dGTP). Heterozygous Rrm1+/Y285A mice exhibited distinct alterations in dNTP pools across various organs, shorter lifespans and earlier tumor onset compared with wild-type controls. Mutational spectrum analysis of tumors revealed two distinct signatures, one resembling a signature extracted from a human cancer harboring a mutation of the same amino acid residue in ribonucleotide reductase, RRM1Y285C. Our findings suggest that mutations in enzymes involved in dNTP metabolism can serve as drivers of cancer development.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Harfe B.D., Jinks-Robertson S.. DNA mismatch repair and genetic instability. Annu. Rev. Genet. 2000; 34:359–399. - PubMed
-
- Rayner E., van Gool I.C., Palles C., Kearsey S.E., Bosse T., Tomlinson I., Church D.N.. A panoply of errors: polymerase proofreading domain mutations in cancer. Nat. Rev. Cancer. 2016; 16:71–81. - PubMed
-
- Venkatesan R.N., Treuting P.M., Fuller E.D., Goldsby R.E., Norwood T.H., Gooley T.A., Ladiges W.C., Preston B.D., Loeb L.A.. Mutation at the polymerase active site of mouse DNA polymerase delta increases genomic instability and accelerates tumorigenesis. Mol. Cell. Biol. 2007; 27:7669–7682. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

