A molecular comparison of [Fe-S] cluster-based homeostasis in Escherichia coli and Pseudomonas aeruginosa
- PMID: 39360836
- PMCID: PMC11559095
- DOI: 10.1128/mbio.01206-24
A molecular comparison of [Fe-S] cluster-based homeostasis in Escherichia coli and Pseudomonas aeruginosa
Abstract
Iron-sulfur [Fe-S] clusters are essential protein cofactors allowing bacteria to perceive environmental redox modification and to adapt to iron limitation. Escherichia coli, which served as a bacterial model, contains two [Fe-S] cluster biogenesis systems, ISC and SUF, which ensure [Fe-S] cluster synthesis under balanced and stress conditions, respectively. However, our recent phylogenomic analyses revealed that most bacteria possess only one [Fe-S] cluster biogenesis system, most often SUF. The opportunist human pathogen Pseudomonas aeruginosa is atypical as it harbors only ISC. Here, we confirmed the essentiality of ISC in P. aeruginosa under both normal and stress conditions. Moreover, P. aeruginosa ISC restored viability, under balanced growth conditions, to an E. coli strain lacking both ISC and SUF. Reciprocally, the E. coli SUF system sustained growth and [Fe-S] cluster-dependent enzyme activities of ISC-deficient P. aeruginosa. Surprisingly, an ISC-deficient P. aeruginosa strain expressing E. coli SUF showed defects in resistance to H2O2 stress and paraquat, a superoxide generator. Similarly, the P. aeruginosa ISC system did not confer stress resistance to a SUF-deficient E. coli mutant. A survey of 120 Pseudomonadales genomes confirmed that all but five species have selected ISC over SUF. While highlighting the great versatility of bacterial [Fe-S] cluster biogenesis systems, this study emphasizes that their contribution to cellular homeostasis must be assessed in the context of each species and its own repertoire of stress adaptation functions. As a matter of fact, despite having only one ISC system, P. aeruginosa shows higher fitness in the face of ROS and iron limitation than E. coli.
Importance: ISC and SUF molecular systems build and transfer Fe-S cluster to cellular apo protein clients. The model Escherichia coli has both ISC and SUF and study of the interplay between the two systems established that the ISC system is the house-keeping one and SUF the stress-responding one. Unexpectedly, our recent phylogenomic analysis revealed that in contrast to E. coli (and related enterobacteria such as Salmonella), most bacteria have only one system, and, in most cases, it is SUF. Pseudomonas aeruginosa fits the general rule of having only one system but stands against the rule by having ISC. This study aims at engineering P. aeruginosa harboring E. coli systems and vice versa. Comparison of the recombinants allowed to assess the functional versatility of each system while appreciating their contribution to cellular homeostasis in different species context.
Keywords: Escherichia coli; Pseudomonas aeruginosa; iron-sulfur biogenesis; stress adaptation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
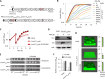

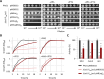

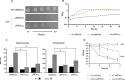
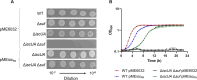


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

