Novel risk loci for COVID-19 hospitalization among admixed American populations
- PMID: 39361370
- PMCID: PMC11449485
- DOI: 10.7554/eLife.93666
Novel risk loci for COVID-19 hospitalization among admixed American populations
Abstract
The genetic basis of severe COVID-19 has been thoroughly studied, and many genetic risk factors shared between populations have been identified. However, reduced sample sizes from non-European groups have limited the discovery of population-specific common risk loci. In this second study nested in the SCOURGE consortium, we conducted a genome-wide association study (GWAS) for COVID-19 hospitalization in admixed Americans, comprising a total of 4702 hospitalized cases recruited by SCOURGE and seven other participating studies in the COVID-19 Host Genetic Initiative. We identified four genome-wide significant associations, two of which constitute novel loci and were first discovered in Latin American populations (BAZ2B and DDIAS). A trans-ethnic meta-analysis revealed another novel cross-population risk locus in CREBBP. Finally, we assessed the performance of a cross-ancestry polygenic risk score in the SCOURGE admixed American cohort. This study constitutes the largest GWAS for COVID-19 hospitalization in admixed Latin Americans conducted to date. This allowed to reveal novel risk loci and emphasize the need of considering the diversity of populations in genomic research.
Keywords: COVID-19; GWAS; SNP; genetics; genomics; none.
© 2024, Diz-de Almeida, Cruz et al.
Conflict of interest statement
SD, RC, AL, JL, Md, IQ, RG, VN, MP, JT, JN, JA, CA, SA, VA, BA, NA, ÁA, EA, CA, MA, MA, RB, MB, MB, JB, MB, LB, MB, RB, MB, EC, CC, LC, JC, RC, MC, JC, MC, MD, AD, Vd, Ad, ED, RE, MF, MF, UF, AF, TF, MG, BG, JG, BÁ, FB, AG, JG, JG, MH, AH, MJ, ML, AB, RL, EM, CM, VM, OM, IM, MM, AM, JM, EM, PM, VC, SO, EO, MP, EP, NS, PP, PP, MP, TP, MP, GP, EP, GP, AP, SL, SR, MF, ER, FR, JR, FR, JR, PR, JS, JS, ET, AT, JT, AT, LT, MU, JV, ZY, RZ, Id, AR, PS, LR, EG, CA, EP, JR, AR, CF, PL, ÁC No competing interests declared
Figures

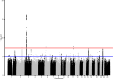
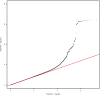

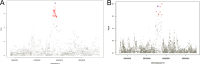
Update of
References
-
- Angeli F, Zappa M, Reboldi G, Gentile G, Trapasso M, Spanevello A, Verdecchia P. The spike effect of acute respiratory syndrome coronavirus 2 and coronavirus disease 2019 vaccines on blood pressure. European Journal of Internal Medicine. 2023;109:12–21. doi: 10.1016/j.ejim.2022.12.004. - DOI - PMC - PubMed
-
- Atkinson EG, Maihofer AX, Kanai M, Martin AR, Karczewski KJ, Santoro ML, Ulirsch JC, Kamatani Y, Okada Y, Finucane HK, Koenen KC, Nievergelt CM, Daly MJ, Neale BM. Tractor uses local ancestry to enable the inclusion of admixed individuals in GWAS and to boost power. Nature Genetics. 2021;53:195–204. doi: 10.1038/s41588-020-00766-y. - DOI - PMC - PubMed
-
- Barbeira AN, Dickinson SP, Bonazzola R, Zheng J, Wheeler HE, Torres JM, Torstenson ES, Shah KP, Garcia T, Edwards TL, Stahl EA, Huckins LM, GTEx Consortium. Nicolae DL, Cox NJ, Im HK. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nature Communications. 2018;9:1825. doi: 10.1038/s41467-018-03621-1. - DOI - PMC - PubMed

