Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study
- PMID: 39361520
- PMCID: PMC11705217
- DOI: 10.1111/febs.17265
Substrate preference, RNA binding and active site versatility of Stenotrophomonas maltophilia nuclease SmNuc1, explained by a structural study
Abstract
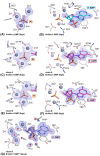
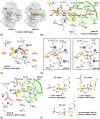
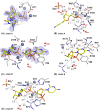
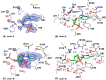
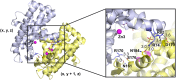
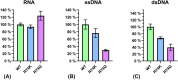
Nucleases of the S1/P1 family have important applications in biotechnology and molecular biology. We have performed structural analyses of SmNuc1 nuclease from Stenotrophomonas maltophilia, including RNA cleavage product binding and mutagenesis in a newly discovered flexible Arg74-motif, involved in substrate binding and product release and likely contributing to the high catalytic rate. The Arg74Gln mutation shifts substrate preference towards RNA. Purine nucleotide binding differs compared to pyrimidines, confirming the plasticity of the active site. The enzyme-product interactions indicate a gradual, stepwise product release. The activity of SmNuc1 towards c-di-GMP in crystal resulted in a distinguished complex with the emerging product 5'-GMP. This enzyme from an opportunistic pathogen relies on specific architecture enabling high performance under broad conditions, attractive for biotechnologies.
Keywords: Stenotrophomonas maltophilia; RNA; S1/P1 nuclease; X‐ray crystallography; c‐di‐GMP cleavage.
© 2024 The Author(s). The FEBS Journal published by John Wiley & Sons Ltd on behalf of Federation of European Biochemical Societies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures








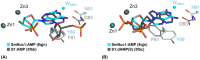




Similar articles
-
A highly active S1-P1 nuclease from the opportunistic pathogen Stenotrophomonas maltophilia cleaves c-di-GMP.FEBS Lett. 2023 Aug;597(16):2103-2118. doi: 10.1002/1873-3468.14683. Epub 2023 Jun 19. FEBS Lett. 2023. PMID: 37309731
-
BsmR degrades c-di-GMP to modulate biofilm formation of nosocomial pathogen Stenotrophomonas maltophilia.Sci Rep. 2017 Jul 5;7(1):4665. doi: 10.1038/s41598-017-04763-w. Sci Rep. 2017. PMID: 28680041 Free PMC article.
-
Insight into the role of substrate-binding residues in conferring substrate specificity for the multifunctional polysaccharide lyase Smlt1473.J Biol Chem. 2014 Jun 27;289(26):18022-32. doi: 10.1074/jbc.M114.571299. Epub 2014 May 7. J Biol Chem. 2014. PMID: 24808176 Free PMC article.
-
Structural and mechanistic determinants of c-di-GMP signalling.Nat Rev Microbiol. 2009 Oct;7(10):724-35. doi: 10.1038/nrmicro2203. Nat Rev Microbiol. 2009. PMID: 19756011 Review.
-
[Progress on the functions of extracellular proteases from Stenotrophomonas maltophilia - A review].Wei Sheng Wu Xue Bao. 2016 May 4;56(5):731-9. Wei Sheng Wu Xue Bao. 2016. PMID: 29727134 Review. Chinese.
References
-
- Ando T (1966) A nuclease specific for heat‐denatured DNA isolated from a product of Aspergillus oryzae . Biochim Biophys Acta 114, 158–168. - PubMed
-
- Mouri T, Hashida W, Shiga I & Teramoto S (1966) Studies on nucleic acid related substances in foodstuff. VII. Nucleic acid degrading enzymes of shiitake (Lentinus edodes). J Ferment Technol 44, 245–258.
-
- Kuninaka A, Otsuka S‐I, Kobayashi Y & Sakaguchi K‐I (1959) Studies on 5′‐phosphodiesterases in microorganisms. Bull Agric Chem Soc Japan 23, 239–243.
-
- Koval T & Dohnalek J (2017) Characteristics and application of S1‐P1 nucleases in biotechnology and medicine. Biotechnol Adv 36, 603–612. - PubMed
-
- Husťakova B, Trundova M, Adamkova K, Koval T, Duskova J & Dohnalek J (2023) A highly active S1‐P1 nuclease from the opportunistic pathogen Stenotrophomonas maltophilia cleaves c‐di‐GMP. FEBS Lett 597, 2103–2118. - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

