Enhancer landscape of lung neuroendocrine tumors reveals regulatory and developmental signatures with potential theranostic implications
- PMID: 39361648
- PMCID: PMC11474083
- DOI: 10.1073/pnas.2405001121
Enhancer landscape of lung neuroendocrine tumors reveals regulatory and developmental signatures with potential theranostic implications
Abstract
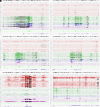
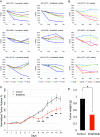
Well-differentiated low-grade lung neuroendocrine tumors (lung carcinoids or LNETs) are histopathologically classified as typical and atypical LNETs, but each subtype is still heterogeneous at both the molecular level and its clinical manifestation. Here, we report genome-wide profiles of primary LNETs' cis-regulatory elements by H3K27ac ChIP-seq with matching RNA-seq profiles. Analysis of these regulatory landscapes revealed three regulatory subtypes, independent of the typical/atypical classification. We identified unique differentiation signals that delineate each subtype. The "proneuronal" subtype emerges under the influence of ASCL1, SOX4, and TCF4 transcription factors, embodying a pronounced proneuronal signature. The "luminal-like" subtype is characterized by gain of acetylation at markers of luminal cells and GATA2 activation and loss of LRP5 and OTP. The "HNF+" subtype is characterized by a robust enhancer landscape driven by HNF1A, HNF4A, and FOXA3, with notable acetylation and expression of FGF signaling genes, especially FGFR3 and FGFR4, pivotal components of the FGF pathway. Our findings not only deepen the understanding of LNETs' regulatory and developmental diversity but also spotlight the HNF+ subtype's reliance on FGFR signaling. We demonstrate that targeting this pathway with FGF inhibitors curtails tumor growth both in vitro and in xenograft models, unveiling a potential vulnerability and paving the way for targeted therapies. Overall, our work provides an important resource for studying LNETs to reveal regulatory networks, differentiation signals, and therapeutically relevant dependencies.
Keywords: FGFR signaling; enhancers; epigenomics; neuroendocrine tumors; pulmonary carcinoids.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures

References
-
- Travis W. D., et al. , Survival analysis of 200 pulmonary neuroendocrine tumors with clarification of criteria for atypical carcinoid and its separation from typical carcinoid. Am. J. Surg. Pathol. 22, 934 (1998). - PubMed
-
- Zhou V. W., Goren A., Bernstein B. E., Charting histone modifications and the functional organization of mammalian genomes. Nat. Rev. Genet. 12, 7–18 (2011). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

