Intestinal Lactobacillus murinus-derived small RNAs target porcine polyamine metabolism
- PMID: 39361652
- PMCID: PMC11474053
- DOI: 10.1073/pnas.2413241121
Intestinal Lactobacillus murinus-derived small RNAs target porcine polyamine metabolism
Abstract
Gut microbiota plays a vital role in host metabolism; however, the influence of gut microbes on polyamine metabolism is unknown. Here, we found germ-free models possess elevated polyamine levels in the colon. Mechanistically, intestinal Lactobacillus murinus-derived small RNAs in extracellular vesicles down-regulate host polyamine metabolism by targeting the expression of enzymes in polyamine metabolism. In addition, Lactobacillus murinus delays recovery of dextran sodium sulfate-induced colitis by reducing polyamine levels in mice. Notably, a decline in the abundance of small RNAs was observed in the colon of mice with colorectal cancer (CRC) and human CRC specimens, accompanied by elevated polyamine levels. Collectively, our study identifies a specific underlying mechanism used by intestinal microbiota to modulate host polyamine metabolism, which provides potential intervention for the treatment of polyamine-associated diseases.
Keywords: CRC; Lactobacillus murinus; colitis; polyamine metabolism; small RNAs.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures
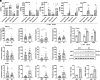

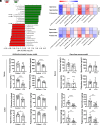

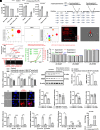

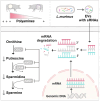
Comment in
-
Gut Microbes Participate in Host Polyamine Metabolism.Proc Natl Acad Sci U S A. 2024 Nov 5;121(45):e2419368121. doi: 10.1073/pnas.2419368121. Epub 2024 Oct 28. Proc Natl Acad Sci U S A. 2024. PMID: 39467143 Free PMC article. No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

