Relocalizing transcriptional kinases to activate apoptosis
- PMID: 39361741
- PMCID: PMC11629774
- DOI: 10.1126/science.adl5361
Relocalizing transcriptional kinases to activate apoptosis
Abstract
Kinases are critical regulators of cellular function that are commonly implicated in the mechanisms underlying disease. Most drugs that target kinases are molecules that inhibit their catalytic activity, but here we used chemically induced proximity to convert kinase inhibitors into activators of therapeutic genes. We synthesized bivalent molecules that link ligands of the transcription factor B cell lymphoma 6 (BCL6) to inhibitors of cyclin-dependent kinases (CDKs). These molecules relocalized CDK9 to BCL6-bound DNA and directed phosphorylation of RNA polymerase II. The resulting expression of pro-apoptotic, BCL6-target genes caused killing of diffuse large B cell lymphoma cells and specific ablation of the BCL6-regulated germinal center response. Genomics and proteomics corroborated a gain-of-function mechanism in which global kinase activity was not inhibited but rather redirected. Thus, kinase inhibitors can be used to context-specifically activate transcription.
Conflict of interest statement
COMPETING INTERESTS:
G.R.C. is a founder and scientific adviser for Foghorn Therapeutics and Shenandoah Therapeutics. N.S.G. is a founder, science advisory board member, and equity holder in Syros, C4, Allorion, Lighthorse, Voronoi, Inception, Matchpoint, CobroVentures, GSK, Shenandoah (board member), Larkspur (board member), and Soltego (board member). T.Z. is a scientific founder, equity holder, and consultant for Matchpoint and an equity holder in Shenandoah. The Gray lab receives or has received research funding from Novartis, Takeda, Astellas, Taiho, Jansen, Kinogen, Arbella, Deerfield, Springworks, Interline, and Sanofi. M.R.G. reports research funding from Sanofi, Kite/Gilead, Abbvie, and Allogene; consulting for Abbvie, Allogene, and Bristol Myers Squibb; honoraria from Tessa Therapeutics, Monte Rosa Therapeutics, and Daiichi Sankyo; and stock ownership of KDAc Therapeutics. Shenandoah has a license from Stanford for the TCIP technology that was invented by G.R.C., S.G., A.K., R.C.S., B.A.K., N.S.G., and T.Z. The remaining authors declare no competing interests.
Figures
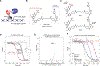


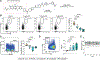
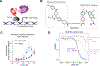
Update of
-
Borrowing Transcriptional Kinases to Activate Apoptosis.bioRxiv [Preprint]. 2023 Oct 25:2023.10.23.563687. doi: 10.1101/2023.10.23.563687. bioRxiv. 2023. Update in: Science. 2024 Oct 4;386(6717):eadl5361. doi: 10.1126/science.adl5361. PMID: 37961702 Free PMC article. Updated. Preprint.
Comment in
-
Bivalent molecules unmute silenced genes.Nat Rev Drug Discov. 2024 Dec;23(12):896. doi: 10.1038/d41573-024-00179-0. Nat Rev Drug Discov. 2024. PMID: 39487217 No abstract available.
References
-
- Wu T et al. , Recent Developments in the Biology and Medicinal Chemistry of CDK9 Inhibitors: An Update. J Med Chem 63, 13228–13257 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

