NEAT1 promotes genome stability via m6A methylation-dependent regulation of CHD4
- PMID: 39362776
- PMCID: PMC11535147
- DOI: 10.1101/gad.351913.124
NEAT1 promotes genome stability via m6A methylation-dependent regulation of CHD4
Abstract
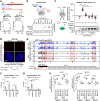
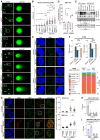
Long noncoding (lnc)RNAs emerge as regulators of genome stability. The nuclear-enriched abundant transcript 1 (NEAT1) is overexpressed in many tumors and is responsive to genotoxic stress. However, the mechanism that links NEAT1 to DNA damage response (DDR) is unclear. Here, we investigate the expression, modification, localization, and structure of NEAT1 in response to DNA double-strand breaks (DSBs). DNA damage increases the levels and N6-methyladenosine (m6A) marks on NEAT1, which promotes alterations in NEAT1 structure, accumulation of hypermethylated NEAT1 at promoter-associated DSBs, and DSB signaling. The depletion of NEAT1 impairs DSB focus formation and elevates DNA damage. The genome-protective role of NEAT1 is mediated by the RNA methyltransferase 3 (METTL3) and involves the release of the chromodomain helicase DNA binding protein 4 (CHD4) from NEAT1 to fine-tune histone acetylation at DSBs. Our data suggest a direct role for NEAT1 in DDR.
Keywords: CHD4; DNA damage response; DNA double-strand breaks; METTL3; NEAT1; long noncoding RNA; paraspeckles.
© 2024 Mamontova et al.; Published by Cold Spring Harbor Laboratory Press.
Figures



References
-
- Adriaens C, Standaert L, Barra J, Latil M, Verfaillie A, Kalev P, Boeckx B, Wijnhoven PW, Radaelli E, Vermi W, et al. 2016. p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat Med 22: 861–868. 10.1038/nm.4135 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
