Serial single-cell RNA sequencing unveils drug resistance and metastatic traits in stage IV breast cancer
- PMID: 39363009
- PMCID: PMC11450160
- DOI: 10.1038/s41698-024-00723-6
Serial single-cell RNA sequencing unveils drug resistance and metastatic traits in stage IV breast cancer
Abstract
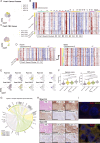
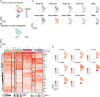
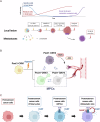
Metastasis is a complex process that remains poorly understood at the molecular levels. We profiled single-cell transcriptomic, genomic, and epigenomic changes associated with cancer cell progression, chemotherapy resistance, and metastasis from a Stage IV breast cancer patient. Pretreatment- and posttreatment-specimens from the primary tumor and distant metastases were collected for single-cell RNA sequencing and subsequent cell clustering, copy number variation (CNV) estimation, transcriptomic factor estimation, and pseudotime analyses. CNV analysis revealed that a small population of pretreatment cancer cells resisted chemotherapy and expanded. New clones including Metastatic Precursor Cells (MPCs), emerged in the posttreatment primary tumors in CNV similar to metastatic cells. MPCs exhibited expression profiles indicative of epithelial-mesenchymal transition. Comparison of MPCs with metastatic cancer cells also revealed dynamic changes in transcription factors and calcitonin pathway gene expression. These findings demonstrate the utility of single-patient clinical sample analysis for understanding tumor drug resistance, regrowth, and metastasis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing Interests.
Figures



References
-
- Parker, J. S. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol.41, 4192–4199 (2023). - PubMed
-
- Kast, K. et al. Impact of breast cancer subtypes and patterns of metastasis on outcome. Breast Cancer Res Treat.150, 621–629 (2015). - PubMed
-
- Tarighati, E., Keivan, H. & Mahani, H. A review of prognostic and predictive biomarkers in breast cancer. Clin. Exp. Med23, 1–16 (2023). - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

