Amine headgroups in ionizable lipids drive immune responses to lipid nanoparticles by binding to the receptors TLR4 and CD1d
- PMID: 39363106
- PMCID: PMC11863198
- DOI: 10.1038/s41551-024-01256-w
Amine headgroups in ionizable lipids drive immune responses to lipid nanoparticles by binding to the receptors TLR4 and CD1d
Abstract
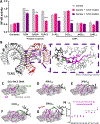
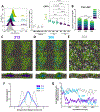
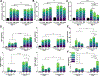
Lipid nanoparticles (LNPs) are the most clinically advanced delivery vehicle for RNA therapeutics, partly because of established lipid structure-activity relationships focused on formulation potency. Yet such knowledge has not extended to LNP immunogenicity. Here we show that the innate and adaptive immune responses elicited by LNPs are linked to their ionizable lipid chemistry. Specifically, we show that the amine headgroups in ionizable lipids drive LNP immunogenicity by binding to Toll-like receptor 4 and CD1d and by promoting lipid-raft formation. Immunogenic LNPs favour a type-1 T-helper-cell-biased immune response marked by increases in the immunoglobulins IgG2c and IgG1 and in the pro-inflammatory cytokines tumour necrosis factor, interferon γ and the interleukins IL-6 and IL-2. Notably, the inflammatory signals originating from these receptors inhibit the production of anti-poly(ethylene glycol) IgM antibodies, preventing the often-observed loss of efficacy in the LNP-mediated delivery of siRNA and mRNA. Moreover, we identified computational methods for the prediction of the structure-dependent innate and adaptive responses of LNPs. Our findings may help accelerate the discovery of well-tolerated ionizable lipids suitable for repeated dosing.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: K.A.W. is an inventor on US patents 9,227,917 (2016) and 9,439,968 (2016) related to the materials described here, and is a consultant for several companies dealing with non-viral RNA delivery.
Figures
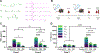


References
-
- Hajj KA & Whitehead KA Tools for translation: non-viral materials for therapeutic mRNA delivery. Nat. Rev. Mater. 2, 17056 (2017).
MeSH terms
Substances
Grants and funding
- DGE1745016/National Science Foundation (NSF)
- F32 EB029345/EB/NIBIB NIH HHS/United States
- F32-EB029345/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- DP2 HD098860/HD/NICHD NIH HHS/United States
- DP2-HD098860/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
LinkOut - more resources
Full Text Sources

