Analysis of codon usage patterns in complete plastomes of four medicinal Polygonatum species (Asparagaceae)
- PMID: 39364010
- PMCID: PMC11447317
- DOI: 10.3389/fgene.2024.1401013
Analysis of codon usage patterns in complete plastomes of four medicinal Polygonatum species (Asparagaceae)
Abstract
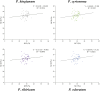
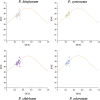
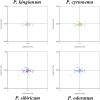
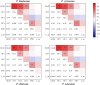
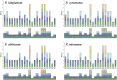
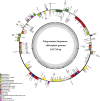
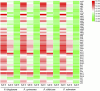
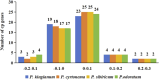
Polygonati Rhizoma and Polygonati odorati Rhizoma, known as "Huangjing" and "Yuzhu" in China, are medicinal Polygonatum species resources with top-grade medical and edible properties. The chloroplast (cp) genome has been used to study species diversity, evolution, and breeding of species for applications in genetic engineering. Codon usage bias (CUB), a common and complex natural phenomenon, is essential for studies of codon optimization of exogenous genes, genetic engineering, and molecular evolution. However, the CUB of medicinal Polygonatum species chloroplast genomes has not been systematically studied. In our study, a detailed analysis of CUB was performed in the medicinal Polygonatum species chloroplast genomes. We investigated the codon bias of 204 plastid protein-coding genes (PCGs) in 4 medicinal Polygonatum species using CodonW and CUSP online software. Through the analysis of the codon bias index, we found that the medicinal Polygonatum species chloroplast genomes had weak codon usage bias. In addition, our results also showed a high preference for AT bases in medicinal Polygonatum species chloroplast genomes, and the preference to use AT-ending codons was observed in these species chloroplast genomes. The neutrality plot, ENC plot, PR2-Bias plot, and correspondence analysis showed that compared with mutation pressure, natural selection was the most important factor of CUB. Based on the comparative analysis of high-frequency codons and high expression codons, we also determined the 10-11 optimal codons of investigative medicinal Polygonatum species. Furthermore, the result of RSCU-based cluster analysis showed that the genetic relationship between different medicinal Polygonatum species could be well reflected. This study provided an essential understanding of CUB and evolution in the medicinal Polygonatum species chloroplast genomes.
Keywords: Polygonatum species; chloroplast genome; codon usage bias; evolution; mutation pressure; natural selection.
Copyright © 2024 Shi, Yuan, Huang and Wen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Comparative Analysis of the Codon Usage Pattern in the Chloroplast Genomes of Gnetales Species.Int J Mol Sci. 2024 Oct 2;25(19):10622. doi: 10.3390/ijms251910622. Int J Mol Sci. 2024. PMID: 39408952 Free PMC article.
-
Comparative Analysis of Codon Bias in the Chloroplast Genomes of Theaceae Species.Front Genet. 2022 Mar 10;13:824610. doi: 10.3389/fgene.2022.824610. eCollection 2022. Front Genet. 2022. PMID: 35360853 Free PMC article.
-
Analysis of the Codon Usage Bias Pattern in the Chloroplast Genomes of Chloranthus Species (Chloranthaceae).Genes (Basel). 2025 Feb 2;16(2):186. doi: 10.3390/genes16020186. Genes (Basel). 2025. PMID: 40004515 Free PMC article.
-
Comparative analysis of codon usage patterns in chloroplast genomes of ten Epimedium species.BMC Genom Data. 2023 Jan 9;24(1):3. doi: 10.1186/s12863-023-01104-x. BMC Genom Data. 2023. PMID: 36624369 Free PMC article.
-
Analysis of codon usage bias of chloroplast genomes in Gynostemma species.Physiol Mol Biol Plants. 2021 Dec;27(12):2727-2737. doi: 10.1007/s12298-021-01105-z. Epub 2021 Dec 1. Physiol Mol Biol Plants. 2021. PMID: 35035132 Free PMC article.
Cited by
-
Comprehensive analysis of the codon usage patterns in the polyprotein coding sequences of the honeybee viruses.Front Vet Sci. 2025 Jul 4;12:1567209. doi: 10.3389/fvets.2025.1567209. eCollection 2025. Front Vet Sci. 2025. PMID: 40687091 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

