Dynamical responses predict a distal site that modulates activity in an antibiotic resistance enzyme
- PMID: 39364073
- PMCID: PMC11443494
- DOI: 10.1039/d4sc03295k
Dynamical responses predict a distal site that modulates activity in an antibiotic resistance enzyme
Abstract
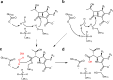
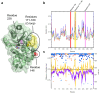
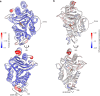
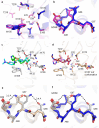
β-Lactamases, which hydrolyse β-lactam antibiotics, are key determinants of antibiotic resistance. Predicting the sites and effects of distal mutations in enzymes is challenging. For β-lactamases, the ability to make such predictions would contribute to understanding activity against, and development of, antibiotics and inhibitors to combat resistance. Here, using dynamical non-equilibrium molecular dynamics (D-NEMD) simulations combined with experiments, we demonstrate that intramolecular communication networks differ in three class A SulpHydryl Variant (SHV)-type β-lactamases. Differences in network architecture and correlated motions link to catalytic efficiency and β-lactam substrate spectrum. Further, the simulations identify a distal residue at position 89 in the clinically important Klebsiella pneumoniae carbapenemase 2 (KPC-2), as a participant in similar networks, suggesting that mutation at this position would modulate enzyme activity. Experimental kinetic, biophysical and structural characterisation of the naturally occurring, but previously biochemically uncharacterised, KPC-2G89D mutant with several antibiotics and inhibitors reveals significant changes in hydrolytic spectrum, specifically reducing activity towards carbapenems without effecting major structural or stability changes. These results show that D-NEMD simulations can predict distal sites where mutation affects enzyme activity. This approach could have broad application in understanding enzyme evolution, and in engineering of natural and de novo enzymes.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures
References
LinkOut - more resources
Full Text Sources

