Transcriptomic insights into pseudorabies virus suppressed cell death pathways in neuroblastoma cells
- PMID: 39364165
- PMCID: PMC11447949
- DOI: 10.3389/fmicb.2024.1430396
Transcriptomic insights into pseudorabies virus suppressed cell death pathways in neuroblastoma cells
Abstract
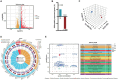
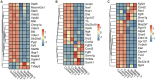
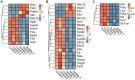
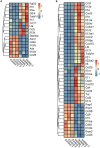
Pseudorabies virus (PRV) exhibits a complex interplay of host-pathogen interactions, primarily by modulating host cell death pathways to optimize its replication and spread in Neuro-2a cells. Using high-throughput RNA sequencing, we identified 2,382 upregulated differentially expressed genes (DEGs) and 3,998 downregulated DEGs, indicating a intricate interaction between viral pathogenesis and host cellular responses. This research offers valuable insights into the molecular processes involved in PRV infection, highlighting the substantial inhibition of crucial cell death pathways in Neuro-2a cells, including necroptosis, pyroptosis, autophagy, ferroptosis, and cuproptosis. Cells infected with PRV exhibit decreased expression of genes critical in these pathways, potentially as a mechanism to avoid host immune reactions and ensure cell survival to support ongoing viral replication. This extensive inhibition of apoptosis and metabolic alterations highlights the sophisticated tactics utilized by PRV, enhancing our comprehension of herpesvirus biology and the feasibility of creating specific antiviral treatments. This research contributes to our understanding of how viruses manipulate host cell death and presents potential opportunities for therapeutic interventions to disrupt the virus's lifecycle.
Keywords: Neuro-2a cells; cell death; inflammatory; pseudorabies virus; transcriptomics.
Copyright © 2024 Cao, Zhang, Zhou and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Pseudorabies virus infected porcine epithelial cell line generates a diverse set of host microRNAs and a special cluster of viral microRNAs.PLoS One. 2012;7(1):e30988. doi: 10.1371/journal.pone.0030988. Epub 2012 Jan 23. PLoS One. 2012. PMID: 22292087 Free PMC article.
-
Pseudorabies Virus Infection Results in a Broad Inhibition of Host Gene Transcription.J Virol. 2022 Jul 13;96(13):e0071422. doi: 10.1128/jvi.00714-22. Epub 2022 Jun 22. J Virol. 2022. PMID: 35730976 Free PMC article.
-
MicroRNA analysis in mouse neuro-2a cells after pseudorabies virus infection.J Neurovirol. 2017 Jun;23(3):430-440. doi: 10.1007/s13365-016-0511-y. Epub 2017 Jan 27. J Neurovirol. 2017. PMID: 28130759
-
A Tug of War: Pseudorabies Virus and Host Antiviral Innate Immunity.Viruses. 2022 Mar 6;14(3):547. doi: 10.3390/v14030547. Viruses. 2022. PMID: 35336954 Free PMC article. Review.
-
Autophagy and Programmed Cell Death Modalities Interplay in HIV Pathogenesis.Cells. 2025 Feb 28;14(5):351. doi: 10.3390/cells14050351. Cells. 2025. PMID: 40072080 Free PMC article. Review.
Cited by
-
Cuproptosis: A Review on Mechanisms, Role in Solid and Hematological Tumors, and Association with Viral Infections.Mediterr J Hematol Infect Dis. 2025 Jul 1;17(1):e2025052. doi: 10.4084/MJHID.2025.052. eCollection 2025. Mediterr J Hematol Infect Dis. 2025. PMID: 40636277 Free PMC article. Review.
-
Lesser-known non-apoptotic programmed cell death in viral infections.Virus Res. 2025 Sep;359:199612. doi: 10.1016/j.virusres.2025.199612. Epub 2025 Aug 7. Virus Res. 2025. PMID: 40759382 Free PMC article. Review.
-
Analysis of differential gene expression in the brain tissue transcriptome of Jiangkou radish piglets infected with porcine pseudorabies virus.SAGE Open Med. 2025 Mar 14;13:20503121251326763. doi: 10.1177/20503121251326763. eCollection 2025. SAGE Open Med. 2025. PMID: 40092414 Free PMC article.
References
-
- Andsberg G., Kokaia Z., Klein R. L., Muzyczka N., Lindvall O., Mandel R. J. (2002). Neuropathological and behavioral consequences of adeno-associated viral vector-mediated continuous intrastriatal neurotrophin delivery in a focal ischemia model in rats. Neurobiol. Dis. 9, 187–204. doi: 10.1006/nbdi.2001.0456 - DOI - PubMed
LinkOut - more resources
Full Text Sources

