Microbial degradation of contaminants of emerging concern: metabolic, genetic and omics insights for enhanced bioremediation
- PMID: 39364263
- PMCID: PMC11446756
- DOI: 10.3389/fbioe.2024.1470522
Microbial degradation of contaminants of emerging concern: metabolic, genetic and omics insights for enhanced bioremediation
Abstract
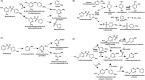
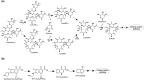
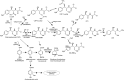
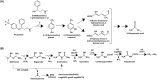
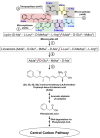
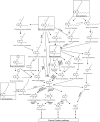
The perpetual release of natural/synthetic pollutants into the environment poses major risks to ecological balance and human health. Amongst these, contaminants of emerging concern (CECs) are characterized by their recent introduction/detection in various niches, thereby causing significant hazards and necessitating their removal. Pharmaceuticals, plasticizers, cyanotoxins and emerging pesticides are major groups of CECs that are highly toxic and found to occur in various compartments of the biosphere. The sources of these compounds can be multipartite including industrial discharge, improper disposal, excretion of unmetabolized residues, eutrophication etc., while their fate and persistence are determined by factors such as physico-chemical properties, environmental conditions, biodegradability and hydrological factors. The resultant exposure of these compounds to microbiota has imposed a selection pressure and resulted in evolution of metabolic pathways for their biotransformation and/or utilization as sole source of carbon and energy. Such microbial degradation phenotype can be exploited to clean-up CECs from the environment, offering a cost-effective and eco-friendly alternative to abiotic methods of removal, thereby mitigating their toxicity. However, efficient bioprocess development for bioremediation strategies requires extensive understanding of individual components such as pathway gene clusters, proteins/enzymes, metabolites and associated regulatory mechanisms. "Omics" and "Meta-omics" techniques aid in providing crucial insights into the complex interactions and functions of these components as well as microbial community, enabling more effective and targeted bioremediation. Aside from natural isolates, metabolic engineering approaches employ the application of genetic engineering to enhance metabolic diversity and degradation rates. The integration of omics data will further aid in developing systemic-level bioremediation and metabolic engineering strategies, thereby optimising the clean-up process. This review describes bacterial catabolic pathways, genetics, and application of omics and metabolic engineering for bioremediation of four major groups of CECs: pharmaceuticals, plasticizers, cyanotoxins, and emerging pesticides.
Keywords: biodegradation; cyanotoxins; metabolic engineering; metabolic pathways; omics; pesticides; pharmaceuticals; plasticizers.
Copyright © 2024 Shah, Malhotra, Papade, Dhamale, Ingale, Kasarlawar and Phale.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Advances in actinobacteria-based bioremediation: mechanistic insights, genetic regulation, and emerging technologies.Biodegradation. 2025 Mar 14;36(2):24. doi: 10.1007/s10532-025-10118-4. Biodegradation. 2025. PMID: 40085365 Review.
-
An innovative approach of bioremediation in enzymatic degradation of xenobiotics.Biotechnol Genet Eng Rev. 2022 Apr;38(1):1-32. doi: 10.1080/02648725.2022.2027628. Epub 2022 Jan 26. Biotechnol Genet Eng Rev. 2022. PMID: 35081881 Review.
-
Sources, pathways, and relative risks of contaminants in surface water and groundwater: a perspective prepared for the Walkerton inquiry.J Toxicol Environ Health A. 2002 Jan 11;65(1):1-142. doi: 10.1080/152873902753338572. J Toxicol Environ Health A. 2002. PMID: 11809004 Review.
-
Microbial Degradation of Naphthalene and Substituted Naphthalenes: Metabolic Diversity and Genomic Insight for Bioremediation.Front Bioeng Biotechnol. 2021 Mar 9;9:602445. doi: 10.3389/fbioe.2021.602445. eCollection 2021. Front Bioeng Biotechnol. 2021. PMID: 33791281 Free PMC article. Review.
-
Omics approaches in bioremediation of environmental contaminants: An integrated approach for environmental safety and sustainability.Environ Res. 2022 Aug;211:113102. doi: 10.1016/j.envres.2022.113102. Epub 2022 Mar 14. Environ Res. 2022. PMID: 35300964 Review.
Cited by
-
Adaptive responses of Gordonia alkanivorans IEGM 1277 to the action of meloxicam and its efficient biodegradation.Front Bioeng Biotechnol. 2025 Jul 23;13:1603975. doi: 10.3389/fbioe.2025.1603975. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40771721 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources

