Reading m6A marks in mRNA: A potent mechanism of gene regulation in plants
- PMID: 39364713
- PMCID: PMC11622538
- DOI: 10.1111/jipb.13781
Reading m6A marks in mRNA: A potent mechanism of gene regulation in plants
Abstract
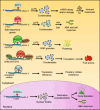
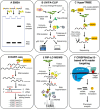
Modifications to RNA have recently been recognized as a pivotal regulator of gene expression in living organisms. More than 170 chemical modifications have been identified in RNAs, with N6-methyladenosine (m6A) being the most abundant modification in eukaryotic mRNAs. The addition and removal of m6A marks are catalyzed by methyltransferases (referred to as "writers") and demethylases (referred to as "erasers"), respectively. In addition, the m6A marks in mRNAs are recognized and interpreted by m6A-binding proteins (referred to as "readers"), which regulate the fate of mRNAs, including stability, splicing, transport, and translation. Therefore, exploring the mechanism underlying the m6A reader-mediated modulation of RNA metabolism is essential for a much deeper understanding of the epigenetic role of RNA modification in plants. Recent discoveries have improved our understanding of the functions of m6A readers in plant growth and development, stress response, and disease resistance. This review highlights the latest developments in m6A reader research, emphasizing the diverse RNA-binding domains crucial for m6A reader function and the biological and cellular roles of m6A readers in the plant response to developmental and environmental signals. Moreover, we propose and discuss the potential future research directions and challenges in identifying novel m6A readers and elucidating the cellular and mechanistic role of m6A readers in plants.
Keywords: RNA metabolism; YTH; epitranscriptomics; m6A modification; m6A reader.
© 2024 The Author(s). Journal of Integrative Plant Biology published by John Wiley & Sons Australia, Ltd on behalf of Institute of Botany, Chinese Academy of Sciences.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
RNA modifications in plant adaptation to abiotic stresses.Plant Commun. 2025 Feb 10;6(2):101229. doi: 10.1016/j.xplc.2024.101229. Epub 2024 Dec 21. Plant Commun. 2025. PMID: 39709520 Free PMC article. Review.
-
Structural Insights into N6-methyladenosine (m6A) Modification in the Transcriptome.Genomics Proteomics Bioinformatics. 2018 Apr;16(2):85-98. doi: 10.1016/j.gpb.2018.03.001. Epub 2018 Apr 27. Genomics Proteomics Bioinformatics. 2018. PMID: 29709557 Free PMC article. Review.
-
A review of m6A modification in plant development and potential quality improvement.Int J Biol Macromol. 2025 May;308(Pt 2):142597. doi: 10.1016/j.ijbiomac.2025.142597. Epub 2025 Mar 27. Int J Biol Macromol. 2025. PMID: 40157682 Review.
-
Function and evolution of RNA N6-methyladenosine modification.Int J Biol Sci. 2020 Apr 15;16(11):1929-1940. doi: 10.7150/ijbs.45231. eCollection 2020. Int J Biol Sci. 2020. PMID: 32398960 Free PMC article. Review.
-
Epitranscriptomic mRNA modifications governing plant stress responses: underlying mechanism and potential application.Plant Biotechnol J. 2022 Dec;20(12):2245-2257. doi: 10.1111/pbi.13913. Epub 2022 Sep 9. Plant Biotechnol J. 2022. PMID: 36002976 Free PMC article. Review.
Cited by
-
RNA modifications in plant adaptation to abiotic stresses.Plant Commun. 2025 Feb 10;6(2):101229. doi: 10.1016/j.xplc.2024.101229. Epub 2024 Dec 21. Plant Commun. 2025. PMID: 39709520 Free PMC article. Review.
-
Epitranscriptomic Control of Drought Tolerance in Rice: The Role of RNA Methylation.Plants (Basel). 2025 Jun 30;14(13):2002. doi: 10.3390/plants14132002. Plants (Basel). 2025. PMID: 40648011 Free PMC article. Review.
-
Research Progress on Natural Products That Regulate miRNAs in the Treatment of Osteosarcoma.Biology (Basel). 2025 Jan 13;14(1):61. doi: 10.3390/biology14010061. Biology (Basel). 2025. PMID: 39857292 Free PMC article. Review.
-
RNA Modification in Metabolism.MedComm (2020). 2025 Mar 10;6(3):e70135. doi: 10.1002/mco2.70135. eCollection 2025 Mar. MedComm (2020). 2025. PMID: 40066222 Free PMC article. Review.
References
-
- Amara, U. , Hu, J. , Cai, J. , and Kang, H. (2023). FLK is an mRNA m6A reader that regulates floral transition by modulating the stability and splicing of FLC in Arabidopsis. Mol. Plant 16: 919–929. - PubMed
-
- Amara, U. , Hu, J. , Park, S.J. , and Kang, H. (2024). ECT12, an YTH‐domain protein, is a potential mRNA m6A reader that affects abiotic stress responses by modulating mRNA stability in Arabidopsis. Plant Physiol. Biochem. 206: 108255. - PubMed
-
- Amara, U. , Shoaib, Y. , and Kang, H. (2022). ALKBH9C, a potential RNA m6A demethylase, regulates the response of Arabidopsis to abiotic stresses and abscisic acid. Plant Cell Environ. 45: 3566–3581. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

