The eATP/P2×7R Axis Drives Quantum Dot-Nanoparticle Induced Neutrophil Recruitment in the Pulmonary Microcirculation
- PMID: 39364760
- PMCID: PMC11615809
- DOI: 10.1002/advs.202404661
The eATP/P2×7R Axis Drives Quantum Dot-Nanoparticle Induced Neutrophil Recruitment in the Pulmonary Microcirculation
Abstract
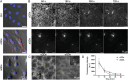
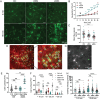
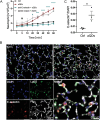
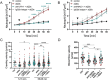
Exposure to nanoparticles (NPs) is frequently associated with adverse cardiovascular effects. In contrast, NPs in nanomedicine hold great promise for precise lung-specific drug delivery, especially considering the extensive pulmonary capillary network that facilitates interactions with bloodstream-suspended particles. Therefore, exact knowledge about effects of engineered NPs within the pulmonary microcirculation are instrumental for future application of this technology in patients. To unravel the real-time dynamics of intravenously delivered NPs and their effects in the pulmonary microvasculature, we employed intravital microscopy of the mouse lung. Only PEG-amine-QDs, but not carboxyl-QDs triggered rapid neutrophil recruitment in microvessels and their subsequent recruitment to the alveolar space and was linked to cellular degranulation, TNF-α, and DAMP release into the circulation, particularly eATP. Stimulation of the ATP-gated receptor P2X7R induced expression of E-selectin on microvascular endothelium thereby mediating the neutrophilic immune response. Leukocyte integrins LFA-1 and MAC-1 facilitated adhesion and decelerated neutrophil crawling on the vascular surface. In summary, this study unravels the complex cascade of neutrophil recruitment during NP-induced sterile inflammation. Thereby we demonstrate novel adverse effects for NPs in the pulmonary microcirculation and provide critical insights for optimizing NP-based drug delivery and therapeutic intervention strategies, to ensure their efficacy and safety in clinical applications.
Keywords: eATP P2X7 axis; innate immune response; intravital microscopy; lung; nanoparticles.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Adhesive Mechanisms of Histone-Induced Neutrophil-Endothelium Interactions in the Muscle Microcirculation.Eur Surg Res. 2016;56(1-2):19-31. doi: 10.1159/000441778. Epub 2015 Nov 18. Eur Surg Res. 2016. PMID: 26575178
-
Microvascular Mechanisms of Polyphosphate-Induced Neutrophil-Endothelial Cell Interactions in vivo.Eur Surg Res. 2019;60(1-2):53-62. doi: 10.1159/000497435. Epub 2019 Mar 25. Eur Surg Res. 2019. PMID: 30909276
-
LPS-induced Lung Platelet Recruitment Occurs Independently from Neutrophils, PSGL-1, and P-Selectin.Am J Respir Cell Mol Biol. 2019 Aug;61(2):232-243. doi: 10.1165/rcmb.2018-0182OC. Am J Respir Cell Mol Biol. 2019. PMID: 30768917 Free PMC article.
-
Immunohistochemical detection of sepsis-induced lung injury in human autopsy material.Leg Med (Tokyo). 2003 Jun;5(2):73-86. doi: 10.1016/s1344-6223(03)00010-5. Leg Med (Tokyo). 2003. PMID: 12935535 Review.
-
Live Intravital Imaging of Cellular Trafficking in the Cardiac Microvasculature-Beating the Odds.Front Immunol. 2019 Nov 26;10:2782. doi: 10.3389/fimmu.2019.02782. eCollection 2019. Front Immunol. 2019. PMID: 31849965 Free PMC article. Review.
References
-
- Buzea C., Pacheco I. I., Robbie K., Biointerphases 2007, 2, MR17. - PubMed
-
- Heires M., New Polit. Econ. 2008, 13, 357.
-
- European Commission , https://eur‐lex.europa.eu/legal‐content/EN/TXT/?uri=CELEX%3A32022H0614%2... (accessed: May 2022).
-
- Oberdörster G., Int. Arch. Occup. Environ. Health 2001, 74, 1. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
