Human and hamster sera correlate well in identifying antigenic drift among SARS-CoV-2 variants, including JN.1
- PMID: 39365051
- PMCID: PMC11578088
- DOI: 10.1128/jvi.00948-24
Human and hamster sera correlate well in identifying antigenic drift among SARS-CoV-2 variants, including JN.1
Abstract
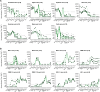
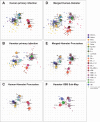
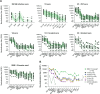
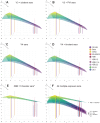
Antigenic assessments of SARS-CoV-2 variants inform decisions to update COVID-19 vaccines. Primary infection sera are often used for assessments, but such sera are rare due to population immunity from SARS-CoV-2 infections and COVID-19 vaccinations. Here, we show that neutralization titers and breadth of matched human and hamster pre-Omicron variant primary infection sera correlate well and generate similar antigenic maps. The hamster antigenic map shows modest antigenic drift among XBB sub-lineage variants, with JN.1 and BA.4/BA.5 variants within the XBB cluster, but with fivefold to sixfold antigenic differences between these variants and XBB.1.5. Compared to sera following only ancestral or bivalent COVID-19 vaccinations, or with post-vaccination infections, XBB.1.5 booster sera had the broadest neutralization against XBB sub-lineage variants, although a fivefold titer difference was still observed between JN.1 and XBB.1.5 variants. These findings suggest that antibody coverage of antigenically divergent JN.1 could be improved with a matched vaccine antigen.IMPORTANCEUpdates to COVID-19 vaccine antigens depend on assessing how much vaccine antigens differ antigenically from newer SARS-CoV-2 variants. Human sera from single variant infections are ideal for discriminating antigenic differences among variants, but such primary infection sera are now rare due to high population immunity. It remains unclear whether sera from experimentally infected animals could substitute for human sera for antigenic assessments. This report shows that neutralization titers of variant-matched human and hamster primary infection sera correlate well and recognize variants similarly, indicating that hamster sera can be a proxy for human sera for antigenic assessments. We further show that human sera following an XBB.1.5 booster vaccine broadly neutralized XBB sub-lineage variants but titers were fivefold lower against the more recent JN.1 variant. These findings support updating the current COVID-19 vaccine variant composition and developing a framework for assessing antigenic differences in future variants using hamster primary infection sera.
Keywords: COVID-19 vaccines; JN.1 variant; SARS-CoV-2 variants; XBB.1.5 booster sera; antigenic assessments; antigenic maps; hamster sera; neutralization titers.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- U.S. Centers for Disease Control and Prevention . 2024. COVID Data Tracker. Available from: https://covid.cdc.gov/covid-data-tracker/#datatracker-home
-
- Pawlowski C, Lenehan P, Puranik A, Agarwal V, Venkatakrishnan AJ, Niesen MJM, O’Horo JC, Virk A, Swift MD, Badley AD, Halamka J, Soundararajan V. 2021. FDA-authorized mRNA COVID-19 vaccines are effective per real-world evidence synthesized across a multi-state health system. Med 2:979–992. doi:10.1016/j.medj.2021.06.007 - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

