Dietary amino acids, macronutrients, vaginal birth, and breastfeeding are associated with the vaginal microbiome in early pregnancy
- PMID: 39365058
- PMCID: PMC11537119
- DOI: 10.1128/spectrum.01130-24
Dietary amino acids, macronutrients, vaginal birth, and breastfeeding are associated with the vaginal microbiome in early pregnancy
Abstract
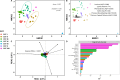
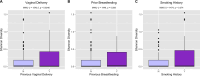
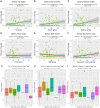
The vaginal microbiome is a key player in the etiology of spontaneous preterm birth. This study aimed to illustrate maternal environmental factors associated with vaginal microbiota composition and function in pregnancy. Women in healthy pregnancy had vaginal microbial sampling from the posterior vaginal fornix performed at 16 weeks gestation. After shotgun metagenomic sequencing, heatmaps of relative abundance data were generated. Community state type (CST) was assigned, and alpha diversity was calculated. Demography, obstetric history, well-being, exercise, and diet using food frequency questionnaires were collected and compared against microbial parameters. A total of 119 pregnant participants had vaginal metagenomic sequencing performed. Factors with strongest association with beta diversity were dietary lysine (adj-R2 0.113, P = 0.002), valine (adj-R2 0.096, P = 0.004), leucine (adj-R2 0.086, P = 0.003), and phenylalanine (adj-R2 0.085, P = 0.005, Fig. 2D). Previous vaginal delivery and breastfeeding were associated with vaginal beta diversity (adj-R2 0.048, P = 0.003; adj-R2 0.045, P = 0.004), accounting for 8.5% of taxonomy variation on redundancy analysis. Dietary fat, starch, and maltose were positively correlated with alpha diversity (fat +0.002 SD/g, P = 0.025; starch +0.002 SD/g, P = 0.043; maltose +0.440 SD/g, P = 0.013), particularly in secretor-positive women. Functional signature was associated with CST, maternal smoking, and dietary phenylalanine, accounting for 8.9%-11% of the variation in vaginal microbiome functional signature. Dietary amino acids, previous vaginal delivery, and breastfeeding history were associated with vaginal beta diversity. Functional signature of the vaginal microbiome differed with community state type, smoking, dietary phenylalanine, and vitamin K. Increased alpha diversity correlated with dietary fat and starch. These data provide a novel snapshot into the associations between maternal environment, nutrition, and the vaginal microbiome.
Importance: This secondary analysis of the MicrobeMom randomized controlled trial reveals that dietary amino acids, macronutrients, previous vaginal birth, and breastfeeding have the strongest associations with vaginal taxonomy in early pregnancy. Function of the vaginal niche is associated mainly by species composition, but smoking, vitamin K, and phenylalanine also play a role. These associations provide an intriguing and novel insight into the association between host factors and diet on the vaginal microbiome in pregnancy and highlight the need for further investigation into the complex interactions between the diet, human gut, and vaginal microbiome.
Keywords: alpha diversity; beta diversity; community state type; diet; environmental; exercise; metagenomic sequencing; microbiota; pregnancy; vagina.
Conflict of interest statement
Authors G.A.C., S.C., R.S., C.F., S.L.K., E.M., F.M.M., R.M., E.O.B., R.O.F., P.M.R., and D.V.S. have no competing interests to declare. P.D.C. is a co-founder, CTO, and shareholder with SeqBiome Ltd., a commercial provider for DNA sequencing and bioinformatics analysis of microbiomes. Research in P.D.C.'s laboratory has been funded by Danone, PepsiCo, and Friesland Campina. P.D.C. has received payment of honoraria from PepsiCo, Abbott, and Yakilt to attend and present at meetings. D.A.M. has received consultancy fees from Freya Biosciences and Kean Health by Psomagen.
Figures


References
-
- Boland K, Bedrani L, Turpin W, Kabakchiev B, Stempak J, Borowski K, Nguyen G, Steinhart AH, Smith MI, Croitoru K, Silverberg MS. 2021. Persistent diarrhea in patients with Crohn’s disease after mucosal healing is associated with lower diversity of the intestinal microbiome and increased dysbiosis. Clin Gastroenterol Hepatol 19:296–304. doi:10.1016/j.cgh.2020.03.044 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

