Genomic evidence of Escherichia coli gut population diversity translocation in leukemia patients
- PMID: 39365076
- PMCID: PMC11520291
- DOI: 10.1128/msphere.00530-24
Genomic evidence of Escherichia coli gut population diversity translocation in leukemia patients
Abstract
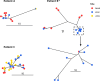
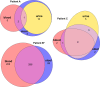
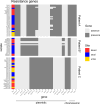
Escherichia coli, a commensal species of the human gut, is an opportunistic pathogen that can reach extra-intestinal compartments, including the bloodstream and the bladder, among others. In non-immunosuppressed patients, purifying or neutral evolution of E. coli populations has been reported in the gut. Conversely, it has been suggested that when migrating to extra-intestinal compartments, E. coli genomes undergo diversifying selection as supported by strong evidence for adaptation. The level of genomic polymorphism and the size of the populations translocating from gut to extra-intestinal compartments is largely unknown. To gain insights into the pathophysiology of these translocations, we investigated the level of polymorphism and the evolutionary forces acting on the genomes of 77 E. coli isolated from various compartments in three immunosuppressed patients. Each patient had a unique strain, which was a mutator in one case. In all instances, we observed that translocation encompasses much of the genomic diversity present in the gut. The same signature of selection, whether purifying or diversifying, and as anticipated, neutral for mutator isolates, was observed in both the gut and bloodstream. Additionally, we found a limited number of non-specific mutations among compartments for non-mutator isolates. In all cases, urine isolates were dominated by neutral selection. These findings indicate that substantial proportions of populations are undergoing translocation and that they present a complex compartment-specific pattern of selection at the patient level.IMPORTANCEIt has been suggested that intra and extra-intestinal compartments differentially constrain the evolution of E. coli strains. Whether host particular conditions, such as immunosuppression, could affect the strain evolutionary trajectories remains understudied. We found that, in immunosuppressed patients, large fractions of E. coli gut populations are translocating with variable modifications of the signature of selection for commensal and pathogenic isolates according to the compartment and/or the patient. Such multiple site sampling should be performed in large cohorts of patients to gain a better understanding of E. coli extra-intestinal diseases.
Keywords: Escherichia coli; adaptation; blood stream infection (BSI); evolution; genomics; immunosuppression; infection; whole-genome sequencing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Tumbarello M, Spanu T, Caira M, Trecarichi EM, Laurenti L, Montuori E, Fianchi L, Leone F, Fadda G, Cauda R, Pagano L. 2009. Factors associated with mortality in bacteremic patients with hematologic malignancies. Diagn Microbiol Infect Dis 64:320–326. doi: 10.1016/j.diagmicrobio.2009.02.008 - DOI - PubMed
-
- Trecarichi EM, Pagano L, Candoni A, Pastore D, Cattaneo C, Fanci R, Nosari A, Caira M, Spadea A, Busca A, Vianelli N, Tumbarello M, HeMABIS Registry—SEIFEM Group, Italy . 2015. Current epidemiology and antimicrobial resistance data for bacterial bloodstream infections in patients with hematologic malignancies: an Italian multicentre prospective survey. Clin Microbiol Infect 21:337–343. doi: 10.1016/j.cmi.2014.11.022 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

