Comprehensive Proteomic Profiling of Human Myocardium Reveals Signaling Pathways Dysregulated in Hypertrophic Cardiomyopathy
- PMID: 39365226
- PMCID: PMC11817648
- DOI: 10.1016/j.jacc.2024.07.043
Comprehensive Proteomic Profiling of Human Myocardium Reveals Signaling Pathways Dysregulated in Hypertrophic Cardiomyopathy
Abstract
Background: Hypertrophic cardiomyopathy (HCM) is the most common genetic cardiac disease. Signaling pathways that link genetic sequence variants to clinically overt HCM and progression to severe forms of HCM remain unknown.
Objectives: The purpose of this study was to identify signaling pathways that are differentially regulated in HCM, using proteomic profiling of human myocardium, confirmed with transcriptomic profiling.
Methods: In this multicenter case-control study, myocardial samples were obtained from cases with HCM and control subjects with nonfailing hearts. Proteomic profiling of 7,289 proteins from myocardial samples was performed using the SomaScan assay (SomaLogic). Pathway analysis of differentially expressed proteins was performed, using a false discovery rate <0.05. Pathway analysis of proteins whose concentrations correlated with clinical indicators of severe HCM (eg, reduced left ventricular ejection fraction, atrial fibrillation, and ventricular tachyarrhythmias) was also executed. Confirmatory analysis of differentially expressed genes was performed using myocardial transcriptomic profiling.
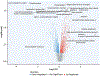
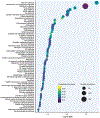
Results: The study included 99 HCM cases and 15 control subjects. Pathway analysis of differentially expressed proteins revealed dysregulation of the Ras-mitogen-activated protein kinase, ubiquitin-mediated proteolysis, angiogenesis-related (eg, hypoxia-inducible factor-1, vascular endothelial growth factor), and Hippo pathways. Pathways known to be dysregulated in HCM, including metabolic, inflammatory, and extracellular matrix pathways, were also dysregulated. Pathway analysis of proteins associated with clinical indicators of severe HCM and of differentially expressed genes supported these findings.
Conclusions: The present study represents the most comprehensive (>7,000 proteins) and largest-scale (n = 99 HCM cases) proteomic profiling of human HCM myocardium to date. Proteomic profiling and confirmatory transcriptomic profiling elucidate dysregulation of both newly recognized (eg, Ras-mitogen-activated protein kinase) and known pathways associated with pathogenesis and progression to severe forms of HCM.
Keywords: hypertrophic cardiomyopathy; myocardium; proteomics; transcriptomics.
Copyright © 2024 American College of Cardiology Foundation. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Funding Support and Author Disclosures This work was supported by the National Institutes of Health (R01 HL157216 and R01 HL168382 to Dr Shimada, UL1 TR001873 to Dr Reilly, K24 HL107643 to Dr Reilly, K24 AG036778 to Dr Maurer, R01 HL170132 to Dr Topkara, R01 HL131872 to Dr Ferrari, and T32 HL007854 to Dr Lumish), the American Heart Association (2 National Clinical and Population Research Awards, 1 Career Development Award, and 1 Transformational Project Award to Dr Shimada), Korea Institute of Oriental Medicine (W22005 to Dr Shimada), Feldstein Medical Foundation (to Dr Shimada), Columbia University Irving Medical Center Precision Medicine Pilot Award (to Dr Shimada), and Columbia University Irving Medical Center Marjorie and Lewis Katz Cardiovascular Research Prize (to Dr Shimada). The funding organizations did not have any role in the study design, collection, analysis, or interpretation of data, in writing of the manuscript, or in the decision to submit the paper for publication. The researchers were independent from the funding organizations. Dr Maurer has received consulting income from Akcea, Alnylam, Eidos Therapeutics, Pfizer, Prothena, Novo Nordisk, and Intellia. Dr Shimada has received research funding from Bristol Myers Squibb; and has received consulting income from Bristol Myers Squibb and Moderna Japan. All other authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Figures



References
-
- Maron BJ. Clinical course and management of hypertrophic cardiomyopathy. N Engl J Med. 2018;379:1977. - PubMed
-
- Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2015;65:1249–1254. - PubMed
-
- Ingles J, Burns C, Barratt A, Semsarian C. Application of genetic testing in hypertrophic cardiomyopathy for preclinical disease detection. Circ Cardiovasc Genet. 2015;8:852–859. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

