Nanoscale organization of cardiac calcium channels is dependent on thyroid hormone status
- PMID: 39365674
- PMCID: PMC11559645
- DOI: 10.1152/ajpheart.00272.2024
Nanoscale organization of cardiac calcium channels is dependent on thyroid hormone status
Abstract
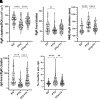
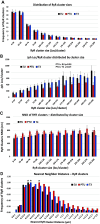
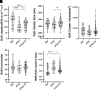
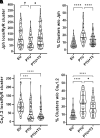
Thyroid hormone dysfunction is frequently observed in patients with chronic illnesses including heart failure, which increases the risk of adverse events. This study examined the effects of thyroid hormones (THs) on cardiac transverse-tubule (TT) integrity, Ca2+ sparks, and nanoscale organization of ion channels in excitation-contraction (EC) coupling, including L-type calcium channel (CaV1.2), ryanodine receptor type 2 (RyR2), and junctophilin-2 (Jph2). TH deficiency was established in adult female rats by propyl-thiouracil (PTU) ingestion for 8 wk; followed by randomization to continued PTU without or with oral triiodo-l-thyronine (T3; 10 µg/kg/day) for an additional 2 wk (PTU + T3). Confocal microscopy of isolated cardiomyocytes (CMs) showed significant misalignment of TTs and increased Ca2+ sparks in thyroid-deficient CMs. Density-based spatial clustering of applications with noise (DBSCAN) analysis of stochastic optical reconstruction microscopy (STORM) images showed decreased (P < 0.0001) RyR2 cluster number per cell area in PTU CMs compared with euthyroid (EU) control myocytes, and this was normalized by T3 treatment. CaV1.2 channels and Jph2 localized within a 210 nm radius of the RyR2 clusters were significantly reduced in PTU myocytes, and these values were increased with T3 treatment. A significant percentage of the RyR2 clusters in the PTU myocytes had neither CaV1.2 nor Jph2, suggesting fewer functional clusters in EC coupling. Nearest neighbor distances between RyR2 clusters were greater (P < 0.001) in PTU cells compared with EU- and T3-treated CMs that correspond to disarray of TTs at the sarcomere z-discs. These results support a regulatory role of T3 in the nanoscale organization of RyR2 clusters and colocalization of CaV1.2 and Jph2 in optimizing EC coupling.NEW & NOTEWORTHY Thyroid hormone (TH) dysfunction exacerbates preexisting heart conditions leading to an increased risk of premature morbidity/mortality. Triiodo-l-thyronine (T3) optimizes cardiac excitation-contraction (EC) coupling by maintaining myocardial T-tubule (TT) structures and organization of calcium ion channels. Single-molecule localization microscopy shows T3 effects on the clustering of ryanodine receptors (RyR2) with colocalization of L-type calcium channels (CaV1.2) and junctophilin-2 (Jph2) at TT-SR structures. Heart disease with subclinical hypothyroidism/low T3 syndrome may benefit from TH treatment.
Keywords: L-type calcium channel; STORM; junctophilin-2; ryanodine receptor-2; thyroid.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures







Comment in
-
Thy and mighty: the importance of T3 thyroid hormone on dyadic structure and function in cardiac health and disease.Am J Physiol Heart Circ Physiol. 2024 Dec 1;327(6):H1384-H1386. doi: 10.1152/ajpheart.00735.2024. Epub 2024 Nov 1. Am J Physiol Heart Circ Physiol. 2024. PMID: 39485299 No abstract available.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

