A novel mean shape based post-processing method for enhancing deep learning lower-limb muscle segmentation accuracy
- PMID: 39365764
- PMCID: PMC11452003
- DOI: 10.1371/journal.pone.0308664
A novel mean shape based post-processing method for enhancing deep learning lower-limb muscle segmentation accuracy
Abstract
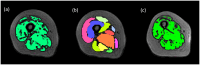
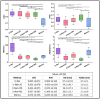
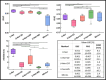
This study aims at improving the lower-limb muscle segmentation accuracy of deep learning approaches based on Magnetic Resonance Imaging (MRI) scans, crucial for the diagnostic and therapeutic processes in musculoskeletal diseases. In general, segmentation methods such as U-Net deep learning neural networks can achieve good Dice Similarity Coefficient (DSC) values, e.g. around 0.83 to 0.91 on various cohorts. Some generic post-processing strategies have been studied to incorporate connectivity constraints into the resulting masks for the purpose of further improving the segmentation accuracy. In this paper, a novel mean shape (MS) based post-processing method is proposed, utilizing Statistical Shape Modelling (SSM) to fine-tune the segmentation output, taking into consideration the muscle anatomical shape. The methodology was compared to existing post-processing techniques and a commercial semi-automatic tool on MRI scans from two cohorts of post-menopausal women (10 Training, 8 Testing, voxel size 1.0x1.0x1.0 mm3). The MS based method obtained a mean DSC of 0.83 across the different analysed muscles and the best performance for the Hausdorff Distance (HD, 20.6 mm) and the Average Symmetric Surface Distance (ASSD, 2.1 mm). These findings highlight the feasibility and potential of using anatomical mean shape in post-processing of human lower-limb muscle segmentation task and indicate that the proposed method can be popularized to other biological organ segmentation mission.
Copyright: © 2024 Lin et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- O. Ronneberger, P. Fischer, and T. Brox, “U-Net: Convolutional Networks for Biomedical Image Segmentation,” arXiv, May 2015. Accessed: Dec. 05, 2023. [Online]. http://arxiv.org/abs/1505.04597
-
- Ni R., Meyer C. H., Blemker S. S., Hart J. M., and Feng X., “Automatic segmentation of all lower limb muscles from high-resolution magnetic resonance imaging using a cascaded three-dimensional deep convolutional neural network,” J. Med. Imaging, vol. 6, no. 04, p. 1, Dec. 2019, doi: 10.1117/1.JMI.6.4.044009 - DOI - PMC - PubMed
-
- Lin Z., Henson W. H., Dowling L., Walsh J., Dall’Ara E., and Guo L., “Automatic segmentation of skeletal muscles from MR images using modified U-Net and a novel data augmentation approach,” Front. Bioeng. Biotechnol., vol. 12, p. 1355735, Feb. 2024, doi: 10.3389/fbioe.2024.1355735 - DOI - PMC - PubMed
-
- Henson W. H., Li X., Lin Z., Guo L., Mazzá C., and Dall’Ara E., “Automatic segmentation of lower limb muscles from MR images of post-menopausal women based on deep learning and data augmentation,” PLOS ONE, vol. 19, no. 4, p. e0299099, Apr. 2024, doi: 10.1371/journal.pone.0299099 - DOI - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

