Toward a CRISPR-based mouse model of Vhl-deficient clear cell kidney cancer: Initial experience and lessons learned
- PMID: 39365820
- PMCID: PMC11474080
- DOI: 10.1073/pnas.2408549121
Toward a CRISPR-based mouse model of Vhl-deficient clear cell kidney cancer: Initial experience and lessons learned
Erratum in
-
Correction to Supporting Information for Stransky et al., Toward a CRISPR-based mouse model of Vhl-deficient clear cell kidney cancer: Initial experience and lessons learned.Proc Natl Acad Sci U S A. 2025 Apr;122(13):e2503473122. doi: 10.1073/pnas.2503473122. Epub 2025 Mar 20. Proc Natl Acad Sci U S A. 2025. PMID: 40112103 Free PMC article. No abstract available.
Abstract
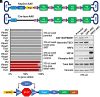
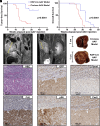
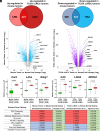
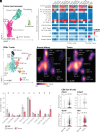
CRISPR is revolutionizing the ability to do somatic gene editing in mice for the purpose of creating new cancer models. Inactivation of the VHL tumor suppressor gene is the signature initiating event in the most common form of kidney cancer, clear cell renal cell carcinoma (ccRCC). Such tumors are usually driven by the excessive HIF2 activity that arises when the VHL gene product, pVHL, is defective. Given the pressing need for a robust immunocompetent mouse model of human ccRCC, we directly injected adenovirus-associated viruses (AAVs) encoding sgRNAs against VHL and other known/suspected ccRCC tumor suppressor genes into the kidneys of C57BL/6 mice under conditions where Cas9 was under the control of one of two different kidney-specific promoters (Cdh16 or Pax8) to induce kidney tumors. An AAV targeting Vhl, Pbrm1, Keap1, and Tsc1 reproducibly caused macroscopic ccRCCs that partially resembled human ccRCC tumors with respect to transcriptome and cell of origin and responded to a ccRCC standard-of-care agent, axitinib. Unfortunately, these tumors, like those produced by earlier genetically engineered mouse ccRCCs, are HIF2 independent.
Keywords: HIF2; PT2399; VHL; ccRCC; mouse models of ccRCC.
Conflict of interest statement
Competing interests statement:W.G.K. has financial interests in Casdin Capital, Cedilla Therapeutics, Circle Pharma, Fibrogen, IconOVir Bio, LifeMine Therapeutics, Lilly Pharmaceuticals, Nextech Invest, and Tango Therapeutics. He also has a royalty interest in the HIF2 inhibitor Belzutifan, which is currently being commercialized by Merck & Co., Inc., Rahway, NJ. L.A.S. has financial interests in Blueprint Medicines. S.S. reports receiving commercial research grants from Bristol-Myers Squibb, AstraZeneca, Exelixis and Novartis; is a consultant/advisory board member for Merck & Co., Inc., Rahway, NJ, AstraZeneca, Bristol-Myers Squibb, CRISPR Therapeutics AG, AACR, and NCI; and receives royalties from Biogenex. E.M.V.A. is a consultant/advisory board member for Enara Bio, Manifold Bio, Monte Rosa, Novartis Institute for Biomedical Research, Serinus Bio; receives research support from Novartis, BMS, Sanofi, NextPoint; reports equity from Tango Therapeutics, Genome Medical, Genomic Life, Enara Bio, Manifold Bio, Microsoft, Monte Rosa, Riva Therapeutics, Syapse, Serinus Bio; intermittent legal consulting on patents for Foaley & Hoag. E.M.P. reports financial interest in Merck & Co., Inc., Rahway, NJ. D.E.L., C.Y., S.D. V.P., and E.M.P. are employees of Merck Pharmaceuticals, which provided the PT2399 used in these studies and which markets the HIF2 inhibitor Belzutifan (Welireg). S.S. reports receiving commercial research grants from Bristol-Myers Squibb, AstraZeneca, Exelixis and Novartis. E.M.V.A. receives research support from Novartis, BMS, Sanofi, NextPoint. Dr. Eliezer M. Van Allen acknowledges shared co-authorship with reviewer Dr. Arul Chinnaiyan on a review in 2020: Mateo et al., Nat. Cancer 2020; 1:1041-1053.
Figures
References
MeSH terms
Substances
Grants and funding
- U01CA236489/HHS | NIH | National Cancer Institute (NCI)
- HHSN261201000031C/CA/NCI NIH HHS/United States
- HHSN261201500001C/CA/NCI NIH HHS/United States
- U01 CA236489/CA/NCI NIH HHS/United States
- HHSN261201500001G/CA/NCI NIH HHS/United States
- HHSN261201500003I/CA/NCI NIH HHS/United States
- T32 CA236754/CA/NCI NIH HHS/United States
- T32CA236754/HHS | NIH | National Cancer Institute (NCI)
- R35CA100068; P50CA101942;/HHS | NIH | National Cancer Institute (NCI)
- P50 CA101942/CA/NCI NIH HHS/United States
- HHSN261201500003C/CA/NCI NIH HHS/United States
- R35 CA210068/CA/NCI NIH HHS/United States
- Contract No.HHSN2612015000031/in part with Federal Funds from the National Cancer Institute, National Institutes of Health
- HHSN261201500001W/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

