SUMOylation of MFF coordinates fission complexes to promote stress-induced mitochondrial fragmentation
- PMID: 39365854
- PMCID: PMC11451547
- DOI: 10.1126/sciadv.adq6223
SUMOylation of MFF coordinates fission complexes to promote stress-induced mitochondrial fragmentation
Abstract
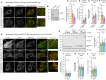
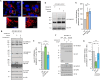
Mitochondria undergo fragmentation in response to bioenergetic stress, mediated by dynamin-related protein 1 (DRP1) recruitment to the mitochondria. The major pro-fission DRP1 receptor is mitochondrial fission factor (MFF), and mitochondrial dynamics proteins of 49 and 51 kilodaltons (MiD49/51), which can sequester inactive DRP1. Together, they form a trimeric DRP1-MiD-MFF complex. Adenosine monophosphate-activated protein kinase (AMPK)-mediated phosphorylation of MFF is necessary for mitochondrial fragmentation, but the molecular mechanisms are unclear. Here, we identify MFF as a target of small ubiquitin-like modifier (SUMO) at Lys151, MFF SUMOylation is enhanced following AMPK-mediated phosphorylation and that MFF SUMOylation regulates the level of MiD binding to MFF. The mitochondrial stressor carbonyl cyanide 3-chlorophenylhydrazone (CCCP) promotes MFF SUMOylation and mitochondrial fragmentation. However, CCCP-induced fragmentation is impaired in MFF-knockout mouse embryonic fibroblasts expressing non-SUMOylatable MFF K151R. These data suggest that the AMPK-MFF SUMOylation axis dynamically controls stress-induced mitochondrial fragmentation by regulating the levels of MiD in trimeric fission complexes.
Figures





References
-
- Detmer S. A., Chan D. C., Functions and dysfunctions of mitochondrial dynamics. Nat. Rev. Mol. Cell Biol. 8, 870–879 (2007). - PubMed
-
- Twig G., Elorza A., Molina A. J. A., Mohamed H., Wikstrom J. D., Walzer G., Stiles L., Haigh S. E., Katz S., Las G., Alroy J., Wu M., Py B. F., Yuan J., Deeney J. T., Corkey B. E., Shirihai O. S., Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27, 433–446 (2008). - PMC - PubMed
-
- Chen H., Chomyn A., Chan D. C., Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J. Biol. Chem. 280, 26185–26192 (2005). - PubMed
-
- Nakada K., Inoue K., Ono T., Isobe K., Ogura A., Goto Y.-I., Nonaka I., Hayashi J.-I., Inter-mitochondrial complementation: Mitochondria-specific system preventing mice from expression of disease phenotypes by mutant mtDNA. Nat. Med. 7, 934–940 (2001). - PubMed
-
- Taguchi N., Ishihara N., Jofuku A., Oka T., Mihara K., Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J. Biol. Chem. 282, 11521–11529 (2007). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

