Site-specific sulfations regulate the physicochemical properties of papillomavirus-heparan sulfate interactions for entry
- PMID: 39365863
- PMCID: PMC11451526
- DOI: 10.1126/sciadv.ado8540
Site-specific sulfations regulate the physicochemical properties of papillomavirus-heparan sulfate interactions for entry
Abstract
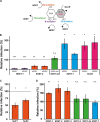
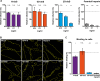
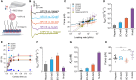
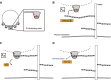
Certain human papillomaviruses (HPVs) are etiological agents for several anogenital and oropharyngeal cancers. During initial infection, HPV16, the most prevalent cancer-causing type, specifically interacts with heparan sulfates (HSs), not only enabling initial cell attachment but also triggering a crucial conformational change in viral capsids termed structural activation. It is unknown, whether these HPV16-HS interactions depend on HS sulfation patterns. Thus, we probed potential roles of HS sulfations using cell-based functional and physicochemical assays, including single-molecule force spectroscopy. Our results demonstrate that N-sulfation of HS is crucial for virus binding and structural activation by providing high-affinity sites, and that additional 6O-sulfation is required to mechanically stabilize the interaction, whereas 2O-sulfation and 3O-sulfation are mostly dispensable. Together, our findings identify the contribution of HS sulfation patterns to HPV16 binding and structural activation and reveal how distinct sulfation groups of HS synergize to facilitate HPV16 entry, which, in turn, likely influences the tropism of HPVs.
Figures

References
-
- J. D. Esko, K. Kimata, U. Lindahl, “Proteoglycans and sulfated glycosaminoglycans” in Essentials of Glycobiology, A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, P. Stanley,C. R. Bertozzi, G. W. Hart, M. E. Etzler, Eds. (Cold Spring Harbor, ed. 2, 2009). - PubMed
-
- Turnbull J., Powell A., Guimond S., Heparan sulfate: Decoding a dynamic multifunctional cell regulator. Trends Cell Biol. 11, 75–82 (2001). - PubMed
-
- Liu J., Thorp S. C., Cell surface heparan sulfate and its roles in assisting viral infections. Med. Res. Rev. 22, 1–25 (2002). - PubMed
-
- Ströh L. J., Stehle T., Glycan engagement by viruses: Receptor switches and specificity. Annu. Rev. Virol. 1, 285 (2014). - PubMed
-
- Esko J. D., Selleck S. B., Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Annu. Rev. Biochem. 71, 435–471 (2002). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

