Maintenance of X chromosome inactivation after T cell activation requires NF-κB signaling
- PMID: 39365876
- PMCID: PMC12088372
- DOI: 10.1126/sciimmunol.ado0398
Maintenance of X chromosome inactivation after T cell activation requires NF-κB signaling
Abstract
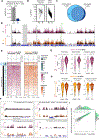
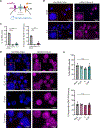
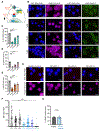
X chromosome inactivation (XCI) balances X-linked gene dosage between sexes. Unstimulated T cells lack cytological enrichment of X-inactive specific transcript (Xist) RNA and heterochromatic modifications on the inactive X chromosome (Xi), which are involved in maintenance of XCI, and these modifications return to the Xi after stimulation. Here, we examined allele-specific gene expression and epigenomic profiles of the Xi in T cells. We found that the Xi in unstimulated T cells is largely dosage compensated and enriched with the repressive H3K27me3 modification but not the H2AK119-ubiquitin (Ub) mark. Upon T cell stimulation mediated by both CD3 and CD28, the Xi accumulated H2AK119-Ub at gene regions of previous H3K27me3 enrichment. T cell receptor (TCR) engagement, specifically NF-κB signaling downstream of the TCR, was required for Xist RNA localization to the Xi. Disruption of NF-κB signaling in mouse and human T cells using genetic deletion, chemical inhibitors, and patients with immunodeficiencies prevented Xist/XIST RNA accumulation at the Xi and altered X-linked gene expression. Our findings reveal a previously undescribed connection between NF-κB signaling pathways, which affects XCI maintenance in T cells in females.
Conflict of interest statement
Competing interests:
The authors declare that they have no competing interests.
Figures




References
-
- Klein SL, Flanagan KL, Sex differences in immune responses. Nat. Rev. Immunol 16, 626–638 (2016). - PubMed
-
- Patin E, Hasan M, Bergstedt J, Rouilly V, Libri V, Urrutia A, Alanio C, Scepanovic P, Hammer C, Jönsson F, Beitz B, Quach H, Lim YW, Hunkapiller J, Zepeda M, Green C, Piasecka B, Leloup C, Rogge L, Huetz F, Peguillet I, Lantz O, Fontes M, Di Santo JP, Thomas S, Fellay J, Duffy D, Quintana-Murci L, Albert ML, M. I. Consortium, Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat. Immunol 19, 302–314 (2018). - PubMed
-
- Melzer S, Zachariae S, Bocsi J, Engel C, Löffler M, Tárnok A, Reference intervals for leukocyte subsets in adults: Results from a population-based study using 10-color flow cytometry. Cytom. B: Clin. Cytom 88, 270–281 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

