Genomic epidemiology of carbapenem-resistant Enterobacterales at a New York City hospital over a 10-year period reveals complex plasmid-clone dynamics and evidence for frequent horizontal transfer of bla KPC
- PMID: 39366703
- PMCID: PMC11610580
- DOI: 10.1101/gr.279355.124
Genomic epidemiology of carbapenem-resistant Enterobacterales at a New York City hospital over a 10-year period reveals complex plasmid-clone dynamics and evidence for frequent horizontal transfer of bla KPC
Abstract
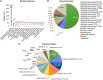
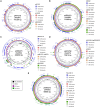
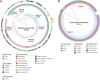
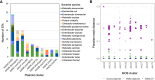
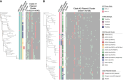
Transmission of carbapenem-resistant Enterobacterales (CRE) in hospitals has been shown to occur through complex, multifarious networks driven by both clonal spread and horizontal transfer mediated by plasmids and other mobile genetic elements. We performed nanopore long-read sequencing on CRE isolates from a large urban hospital system to determine the overall contribution of plasmids to CRE transmission and identify specific plasmids implicated in the spread of bla KPC (the Klebsiella pneumoniae carbapenemase [KPC] gene). Six hundred and five CRE isolates collected between 2009 and 2018 first underwent Illumina sequencing for genome-wide genotyping; 435 bla KPC-positive isolates were then successfully nanopore sequenced to generate hybrid assemblies including circularized bla KPC-harboring plasmids. Phylogenetic analysis and Mash clustering were used to define putative clonal and plasmid transmission clusters, respectively. Overall, CRE isolates belonged to 96 multilocus sequence types (STs) encoding bla KPC on 447 plasmids which formed 54 plasmid clusters. We found evidence for clonal transmission in 66% of CRE isolates, over half of which belonged to four clades comprising K. pneumoniae ST258. Plasmid-mediated acquisition of bla KPC occurred in 23%-27% of isolates. While most plasmid clusters were small, several plasmids were identified in multiple different species and STs, including a highly promiscuous IncN plasmid and an IncF plasmid putatively spreading bla KPC from ST258 to other clones. Overall, this points to both the continued dominance of K. pneumoniae ST258 and the dissemination of bla KPC across clones and species by diverse plasmid backbones. These findings support integrating long-read sequencing into genomic surveillance approaches to detect the hitherto silent spread of carbapenem resistance driven by mobile plasmids.
© 2024 Gomez-Simmonds et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
Molecular epidemiology of carbapenem resistant Enterobacteriaceae in Valle d'Aosta region, Italy, shows the emergence of KPC-2 producing Klebsiella pneumoniae clonal complex 101 (ST101 and ST1789).BMC Microbiol. 2015 Nov 9;15(1):260. doi: 10.1186/s12866-015-0597-z. BMC Microbiol. 2015. PMID: 26552763 Free PMC article.
-
Molecular dissection of an outbreak of carbapenem-resistant enterobacteriaceae reveals Intergenus KPC carbapenemase transmission through a promiscuous plasmid.mBio. 2011 Nov 1;2(6):e00204-11. doi: 10.1128/mBio.00204-11. Print 2011. mBio. 2011. PMID: 22045989 Free PMC article.
-
Genomic Epidemiology of Complex, Multispecies, Plasmid-Borne blaKPC Carbapenemase in Enterobacterales in the United Kingdom from 2009 to 2014.Antimicrob Agents Chemother. 2020 Apr 21;64(5):e02244-19. doi: 10.1128/AAC.02244-19. Print 2020 Apr 21. Antimicrob Agents Chemother. 2020. PMID: 32094139 Free PMC article.
-
Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding.Trends Microbiol. 2014 Dec;22(12):686-96. doi: 10.1016/j.tim.2014.09.003. Epub 2014 Oct 7. Trends Microbiol. 2014. PMID: 25304194 Free PMC article. Review.
-
Rapidly spreading Enterobacterales with OXA-48-like carbapenemases.J Clin Microbiol. 2025 Feb 19;63(2):e0151524. doi: 10.1128/jcm.01515-24. Epub 2025 Jan 6. J Clin Microbiol. 2025. PMID: 39760498 Free PMC article. Review.
References
-
- Annavajhala MK, Gomez-Simmonds A, Macesic N, Sullivan SB, Kress A, Khan SD, Giddins MJ, Stump S, Kim GI, Narain R, et al. 2019. Colonizing multidrug-resistant bacteria and the longitudinal evolution of the intestinal microbiome after liver transplantation. Nat Commun 10: 4715. 10.1038/s41467-019-12633-4 - DOI - PMC - PubMed
-
- Bowers JR, Kitchel B, Driebe EM, MacCannell DR, Roe C, Lemmer D, de Man T, Rasheed JK, Engelthaler DM, Keim P, et al. 2015. Genomic analysis of the emergence and rapid global dissemination of the clonal group 258 Klebsiella pneumoniae pandemic. PLoS ONE 10: e0133727. 10.1371/journal.pone.0133727 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
