SLMAP3 is essential for neurulation through mechanisms involving cytoskeletal elements, ABP, and PCP
- PMID: 39366759
- PMCID: PMC11452652
- DOI: 10.26508/lsa.202302545
SLMAP3 is essential for neurulation through mechanisms involving cytoskeletal elements, ABP, and PCP
Abstract
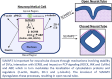
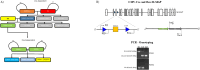
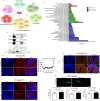
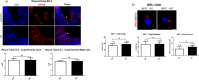
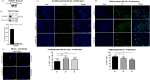
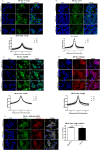
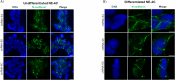
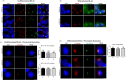
SLMAP3 is a tail-anchored membrane protein that targets subcellular organelles and is believed to regulate Hippo signaling. The global loss of SLMAP3 causes late embryonic lethality in mice, with some embryos exhibiting neural tube defects such as craniorachischisis. We show here that SLMAP3 -/- embryos display reduced length and increased width of neural plates, signifying arrested convergent extension. The expression of planar cell polarity (PCP) components Dvl2/3 and the activity of the downstream targets ROCK2, cofilin, and JNK1/2 were dysregulated in SLMAP3 -/- E12.5 brains. Furthermore, the cytoskeletal proteins (γ-tubulin, actin, and nestin) and apical components (PKCζ and ZO-1) were mislocalized in neural tubes of SLMAP3 -/- embryos, with a subsequent decrease in colocalization of PCP proteins (Fzd6 and pDvl2). However, no changes in PCP or cytoskeleton proteins were found in cultured neuroepithelial cells depleted of SLMAP3, suggesting an essential requirement for SLMAP3 for these processes in vivo for neurulation. The loss of SLMAP3 had no impact on Hippo signaling in SLMAP3 -/- embryos, brains, and neural tubes. Proteomic analysis revealed SLMAP3 in an interactome with cytoskeletal components, including nestin, tropomyosin 4, intermediate filaments, plectin, the PCP protein SCRIB, and STRIPAK members in embryonic brains. These results reveal a crucial role of SLMAP3 in neural tube development by regulating the cytoskeleton organization and PCP pathway.
© 2024 Rehmani et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
-
- Wigle JT, Eisenstat DD (2023) Neural induction and regionalization Neurodevelopmental Pediatrics. New York, NY: Springer International Publishing: 29–38.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
