Mitochondria remodeling during endometrial stromal cell decidualization
- PMID: 39366760
- PMCID: PMC11452479
- DOI: 10.26508/lsa.202402627
Mitochondria remodeling during endometrial stromal cell decidualization
Abstract
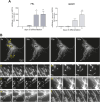
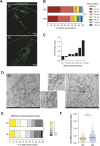
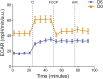
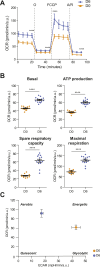
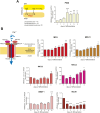
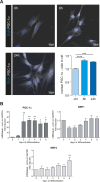
Upon hormonal stimulation, uterine endometrial stromal cells undergo a dramatic morpho-functional metamorphosis that allows them to secrete large amounts of matrix proteins, cytokines, and growth factors. This step, known as decidualization, is crucial for embryo implantation. We previously demonstrated how the secretory pathway is remodelled during this process. Here we show that hormonal stimulation rapidly induces the expression of many mitochondrial genes, encoded in both the mitochondrial and the nuclear genomes. Altogether, the mitochondrial network quadruples its size and establishes more contacts with the ER. This new organization results in the increased respiratory capacity of decidualized cells. These findings reveal how achieving an efficient secretory phenotype requires a radical metabolic rewiring.
© 2024 Dalla Torre et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
