MDM2 induces pro-inflammatory and glycolytic responses in M1 macrophages by integrating iNOS-nitric oxide and HIF-1α pathways in mice
- PMID: 39366973
- PMCID: PMC11452520
- DOI: 10.1038/s41467-024-53006-w
MDM2 induces pro-inflammatory and glycolytic responses in M1 macrophages by integrating iNOS-nitric oxide and HIF-1α pathways in mice
Abstract
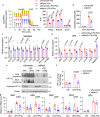
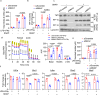
M1 macrophages induce protective immunity against infection, but also contribute to metabolic and inflammatory diseases. Here we show that the E3 ubiquitin ligase, MDM2, promotes the glycolytic and inflammatory activities of M1 macrophage by increasing the production of IL-1β, MCP-1 and nitric oxide (NO). Mechanistically, MDM2 triggers the ubiquitination and degradation of E3 ligase, SPSB2, to stabilize iNOS and increases production of NO, which s-nitrosylates and activates HIF-1α for triggering the glycolytic and pro-inflammatory programs in M1 macrophages. Myeloid-specific haplodeletion of MDM2 in mice not only blunts LPS-induced endotoxemia and NO production, but also alleviates obesity-induced adipose tissue-resident macrophage inflammation. By contrast, MDM2 haplodeletion induces higher mortality, tissue damage and bacterial burden, and also suppresses M1 macrophage response, in the cecal ligation and puncture-induced sepsis mouse model. Our findings thus identify MDM2 as an activator of glycolytic and inflammatory responses in M1 macrophages by connecting the iNOS-NO and HIF-1α pathways.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures








References
-
- Benoit, M., Desnues, B. & Mege, J. L. Macrophage polarization in bacterial infections. J. Immunol.181, 3733–3739 (2008). - PubMed
-
- Shapouri-Moghaddam, A. et al. Macrophage plasticity, polarization, and function in health and disease. J. Cell Physiol.233, 6425–6440 (2018). - PubMed
-
- McGettrick, A. F. & O’Neill, L. A. J. The role of HIF in immunity and inflammation. Cell Metab.32, 524–536 (2020). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

